华大stereo-seq 添加上底片转换成10X格式方便scanpy和seurat读入分析
华大stereo-seq 添加上底片转换成10X格式方便scanpy和seurat读入分析
示例数据: https://en.stomics.tech/col1241/index.html
0.SAW流程:
saw count \
--id=Demo_Mouse_Brain \
--sn=C04042E3 \
--output ./ \
--omics=transcriptomics \
--kit-version="Stereo-seq T FF V1.3" \
--sequencing-type="PE75_50+100" \
--chip-mask=./C04042E3/fastq/C04042E3.barcodeToPos.h5 \
--organism=mouse \
--tissue=brain \
--threads-num 20 \
--fastqs./C04042E3/fastq \
--reference=/share/ref/Mus_musculus/Mus_musculus_index \
--image-tar=./C04042E3/fastq/C04042E3_SC_20240925_142342_4.1.0.tar.gz
1.将SAW跑出来的gef文件转换成gem文件并压缩:
#tissue转换
saw convert --threads-num 20 gef2gem --gef ../01.saw/Demo_Mouse_Brain/outs/feature_expression/C04042E3.tissue.gef \
--gem C04042E3.tissue.gem --bin-size 1
gzip C04042E3.tissue.gem
#cellbin转换
saw convert --threads-num 20 gef2gem --gef ../01.saw/Demo_Mouse_Brain/outs/feature_expression/C04042E3.tissue.gef \
--cellbin-gef ../01.saw/Demo_Mouse_Brain/outs/feature_expression/C04042E3.adjusted.cellbin.gef \
--cellbin-gem C04042E3.adjusted.cellbin.gem --bin-size 1
gzip C04042E3.adjusted.cellbin.gem
2.使用R代码转换成10X结果:
可以通过参数设置图片不同的分辨率大小,binsize
Rscript $scripts//STOTo10X/gemTo10X.r \
--gemf C04042E3.tissue.gem.gz \
-t ../01.saw/Demo_Mouse_Brain/outs/image/C04042E3_HE_regist.tif \
--bin 100 --lowres 0.05 --hires 0.1 --slide_name C04042E3 --outdir bin100
Rscript $scripts//STOTo10X/gemTo10X.r \
--gemf C04042E3.tissue.gem.gz \
-t ../01.saw/Demo_Mouse_Brain/outs/image/C04042E3_HE_regist.tif \
--bin 50 --lowres 0.05 --hires 0.1 --slide_name C04042E3 --outdir bin50
Rscript $scripts//STOTo10X/gemTo10X.r \
--gemf C04042E3.adjusted.cellbin.gem.gz \
-t ../01.saw/Demo_Mouse_Brain/outs/image/C04042E3_HE_regist.tif \
--bin 1 --lowres 0.05 --hires 0.1 --slide_name C04042E3 --outdir cellbin --cellbin
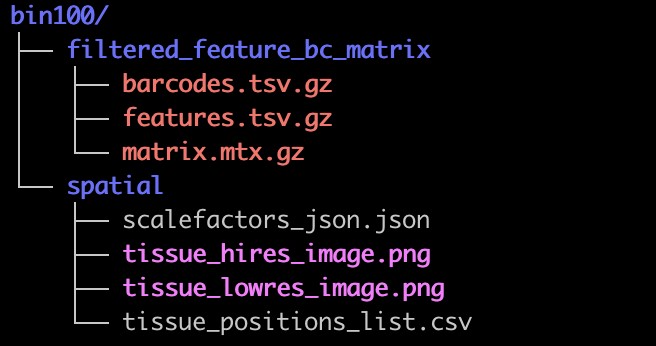
3.结果目录展示:
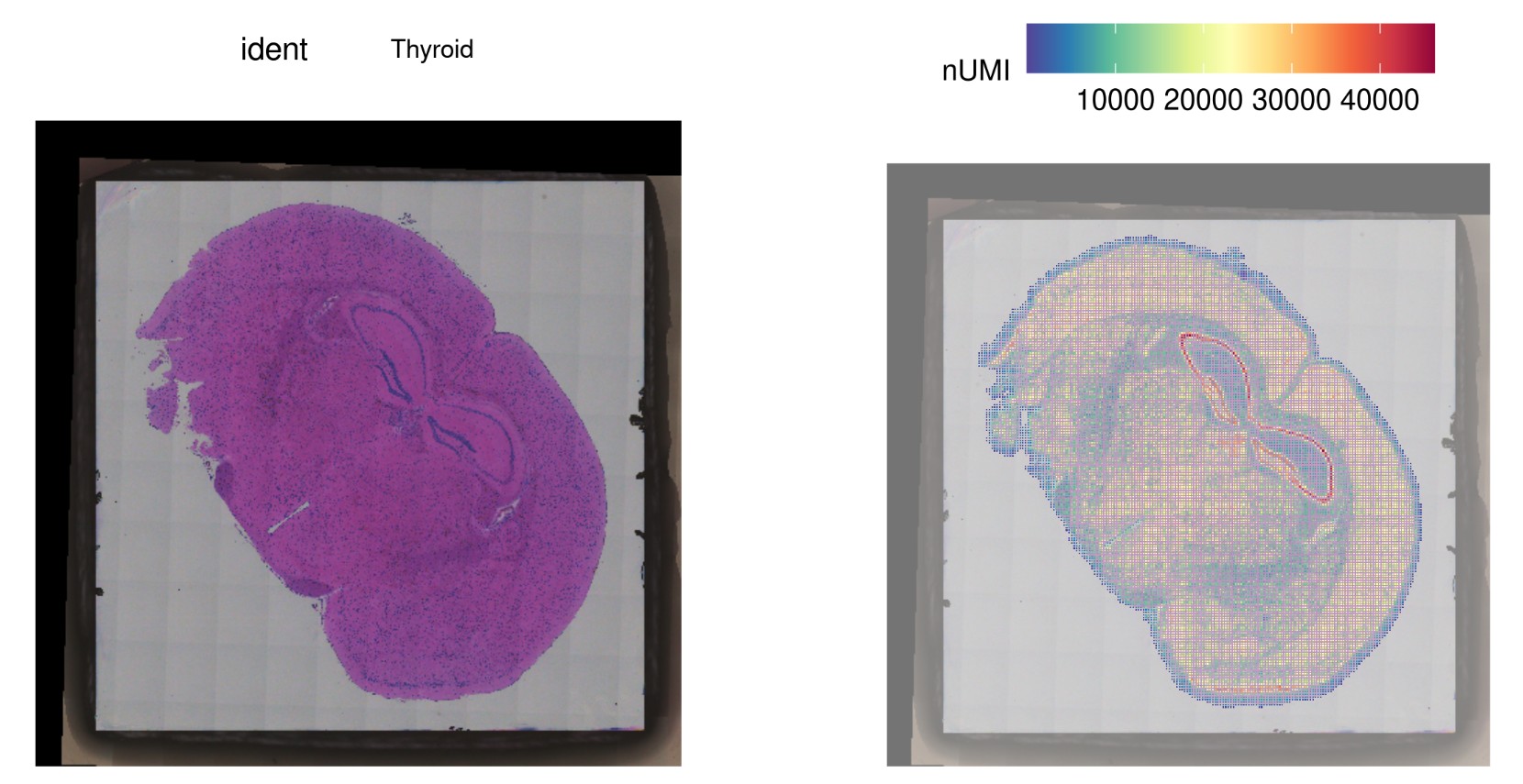
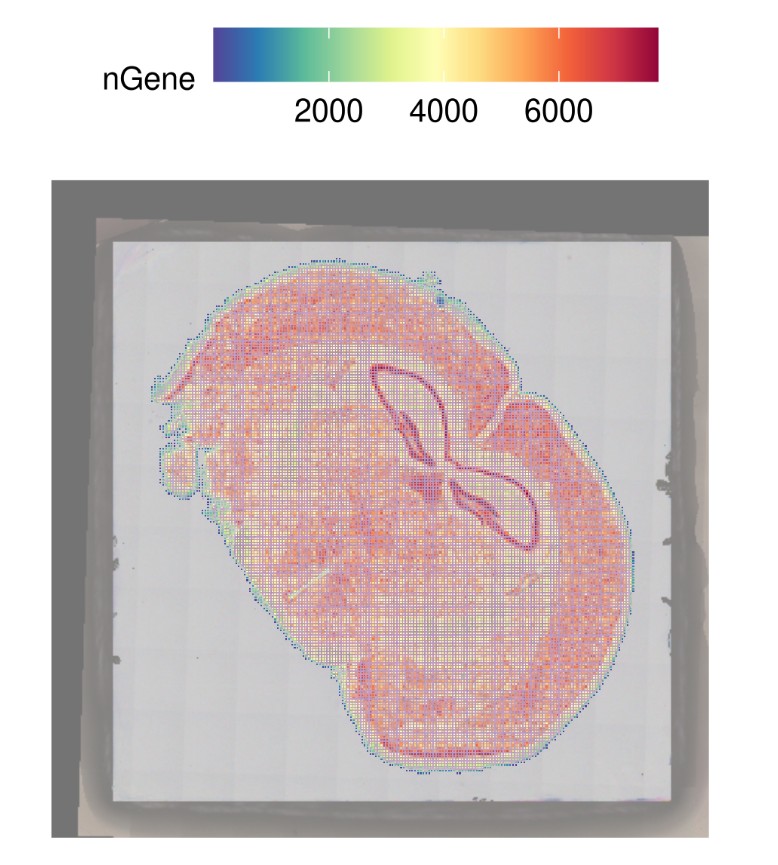
4.R 读入绘制结果图:
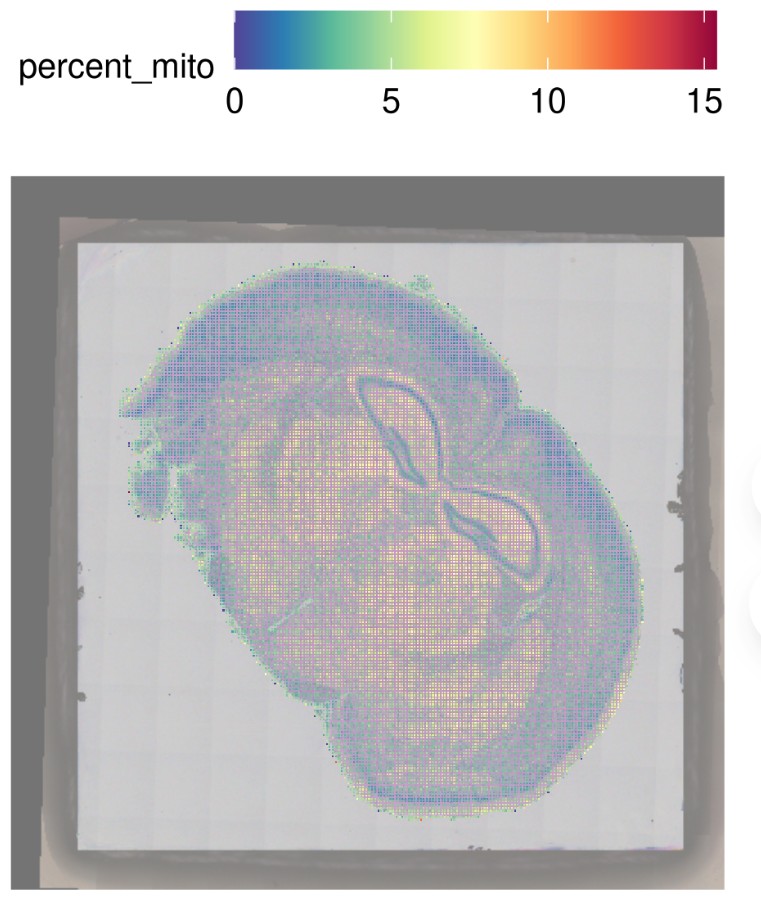
scanpy读入分析结果:
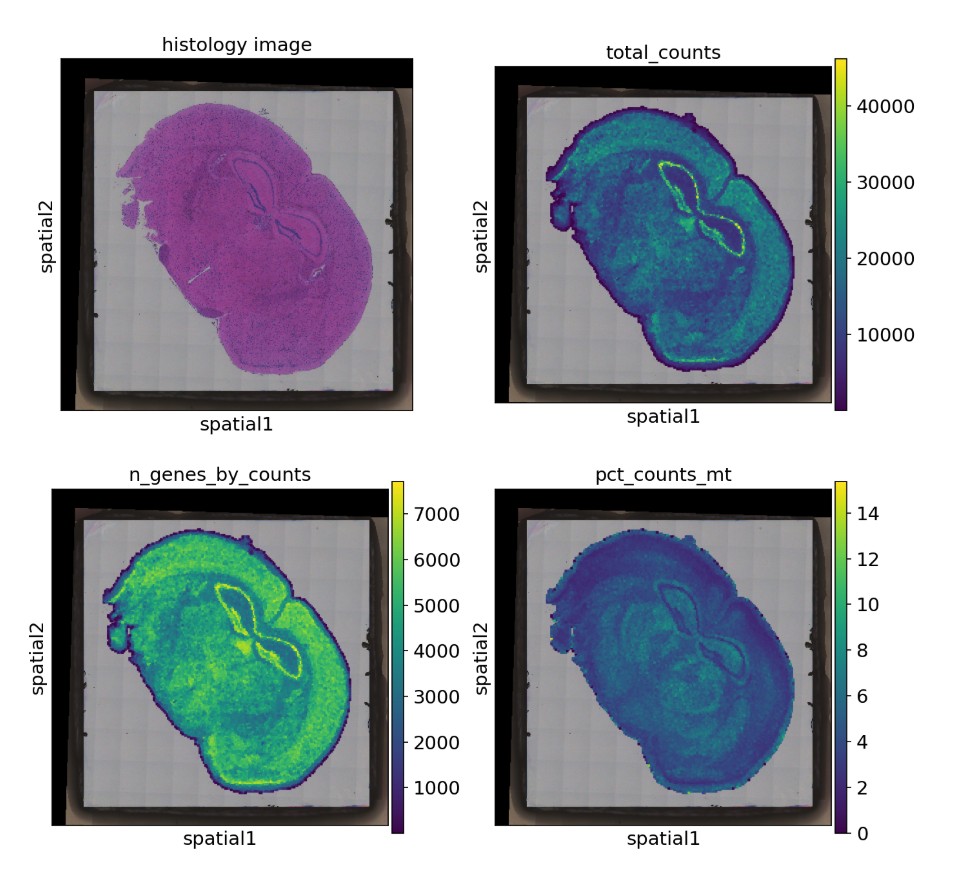
- 发表于 2025-06-24 15:27
- 阅读 ( 2887 )
- 分类:转录组
你可能感兴趣的文章
- 空间转录组细胞分割 312 浏览
- SAW 空间转录组参考基因组制作 2144 浏览
- SAW华大时空软件使用 2441 浏览
- 空间转录组实操课程 3128 浏览
- 空间转录组动力学分析 2183 浏览
2 条评论
请先 登录 后评论
