快速生成iTOL配置文件美化你的进化树
快速生成iTOL配置文件美化你的进化树
更好的阅读体验: https://mp.weixin.qq.com/s/jdBpM_cFBZLZtjtHUTl-8w
iTOL(https://itol.embl.de/)是一款很好的进化树美化软件,但是他的配置文件制作非常麻烦,给大家的使用带来了不便,这里介绍一个工具帮助大家可以从表格文件中快速生成itol需要的配置文件美化进化树。下面给出例子:
. 示例数据
1. 进化树数据 abunt-tree.nwk:
((OTU_47:0.101892516,OTU_46:0.205918075)0.999:0.12637554249999997,((OTU_7:0.079452354,(OTU_9:0.03389913,OTU_35:0.011411273)0.857:0.016187755)0.936:0.055036757,(((OTU_10:0.178702248,(OTU_27:0.105807393,(OTU_21:0.021742283,OTU_23:0.036247052)1.000:0.119621254)0.934:0.046734483)0.847:0.024928304,(OTU_359:0.115728596,((OTU_6:0.017083581,OTU_13:0.043230986)0.991:0.04686027,(OTU_5:0.048441113,OTU_17:0.040966445)0.874:0.016500727)0.998:0.076649272)0.884:0.025403911)0.896:0.031188452,((((OTU_4:0.016277276,OTU_1049:0.018764271)1.000:0.10486842,(OTU_61:0.033971226,OTU_66:0.014225781)0.949:0.029591816)0.938:0.030200498,(OTU_50:0.039428411,((OTU_14:0.027067745,OTU_24:0.006239384)0.713:0.003838445,(OTU_28:0.035593371,(OTU_12:0.019817974,OTU_1263:0.010126282)1.000:0.051942928)0.889:0.016154455)1.000:0.114060029)0.353:0.019135966)0.931:0.027767722,((OTU_26:0.011320021,(OTU_2:0.013893208,OTU_509:0.013184741)0.420:0.004818858)0.869:0.012841976,(((OTU_20:0.014868571,OTU_16:0.015213271)0.912:0.014485178,(OTU_19:0.042730937,OTU_31:0.03306436)0.071:0.011603998)0.445:0.011922646,(OTU_107:0.057678909,(OTU_8:5e-09,(OTU_3:0.01725298,((OTU_18:0.014018692,(OTU_11:0.009416427,OTU_756:0.019954543)0.815:0.007464513)0.616:0.003595574,(OTU_41:0.01321098,OTU_601:0.020022049)0.096:0.008198644)0.883:0.015142165)0.979:0.01496564)0.999:0.044655374)0.971:0.028869096)0.864:0.008905959)0.995:0.05488604)0.835:0.02976759)0.883:0.039831577):0.013815376500000032)root;
2. 数据data(abunt-metadata.txt):
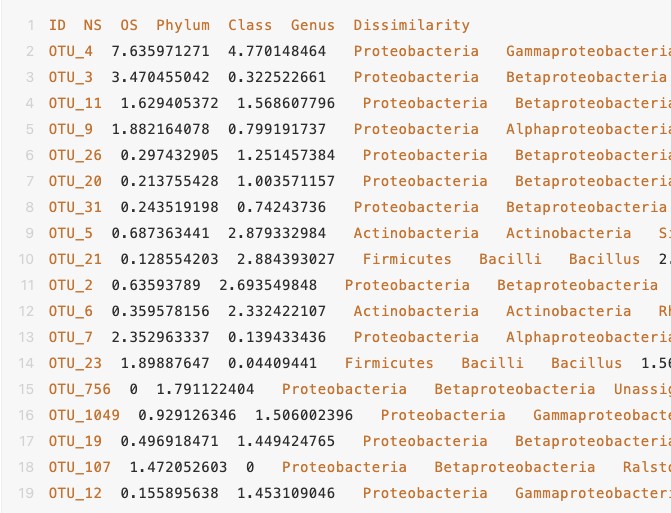
. 代码示例
用到的工具为一个R包,可以读入表格快速,然后将生成好的配置文件,在iTOL网站上拖拽过去即可:
devtools::install_github("TongZhou2017/itol.toolkit")
library(itol.toolkit)
library(data.table)
setwd("/phylo_tree")
tree_1 <- "abunt-tree.nwk"
hub_1 <- create_hub(tree_1)
data_file_1<- "abunt-metadata.txt"
data_1 <- data.table::fread(data_file_1)
# relabel by genus
unit_1 <- create_unit(data = data_1 %>% select(ID, Genus),
key = "itol_3al_1_labels",
type = "LABELS",
tree = tree_1)
write_unit(unit_1,paste0(getwd(),"/itol_3al_1_labels_Genus.txt"))
# tree_colors range by phylum 按照门填充颜色
unit_2 <- create_unit(data = data_1 %>% select(ID, Phylum),
key = "itol_3al_2_range",
type = "TREE_COLORS",
subtype = "range",
tree = tree_1)
write_unit(unit_2,paste0(getwd(),"/itol_3al_2_range_Phylum.txt"))
# color_strip by class 分组圆环
set.seed(123)
unit_3 <- create_unit(data = data_1 %>% select(ID, Class),
key = "itol_3al_3_strip",
type = "DATASET_COLORSTRIP",
color = "wesanderson",
tree = tree_1)
unit_3@common_themes$basic_theme$margin <- 50
write_unit(unit_3,paste0(getwd(),"/itol_3al_3_strip_Class.txt"))
# simple_bar by NS
unit_4 <- create_unit(data = data_1 %>% select(ID, NS),
key = "itol_3al_4_simplebar",
type = "DATASET_SIMPLEBAR",
tree = tree_1)
unit_4@specific_themes$basic_plot$size_max <- 100
write_unit(unit_4,paste0(getwd(),"/itol_3al_4_simplebar.txt"))
# simple_bar by OS
unit_5 <- create_unit(data = data_1 %>% select(ID, NS,OS),
key = "itol_3al_5_multibar",
type = "DATASET_MULTIBAR",
tree = tree_1)
unit_5@specific_themes$basic_plot$size_max <- 100
write_unit(unit_5,paste0(getwd(),"/itol_3al_5_multibar.txt"))
# simple_bar by OS
unit_6 <- create_unit(data = data_1 %>% select(ID, Dissimilarity),
key = "itol_3al_6_gradient",
type = "DATASET_GRADIENT",
tree = tree_1)
unit_6@specific_themes$heatmap$color$min <- "#0000ff"
unit_6@specific_themes$heatmap$color$max <- "#ff0000"
write_unit(unit_6,paste0(getwd(),"/itol_3al_6_gradient.txt"))
unit_7 <- create_unit(data = data_1 %>% select(ID, NS,OS),
key = "itol_7_heatmap",
type = "DATASET_HEATMAP",
tree = tree_1)
write_unit(unit_7,paste0(getwd(),"/itol_7_heatmap.txt"))
最后的效果:
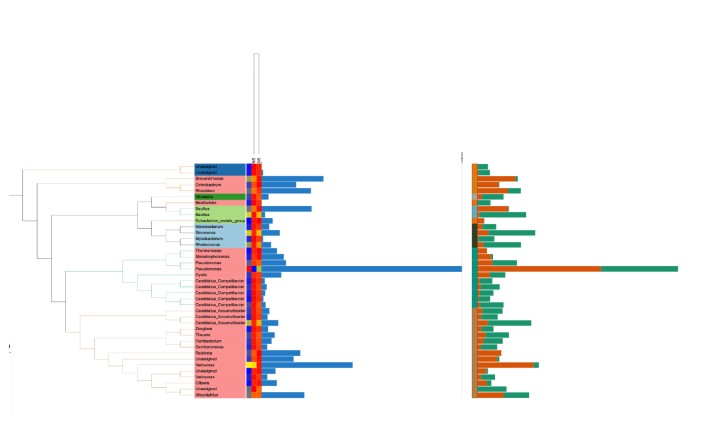
- 发表于 2023-11-08 10:29
- 阅读 ( 3648 )
- 分类:R
你可能感兴趣的文章
- 带推断线的进化树 308 浏览
- 给进化树标定化石时间 4620 浏览
- itol添加圆环分组和节点标签时位置调整 6395 浏览
- 如何用R语言绘制进化树+热图 11716 浏览
- 进化树构建的方法及原理详解 49321 浏览
0 条评论
请先 登录 后评论
