SCP 安装成功的单细胞转录组 docker镜像使用 single-cell-v5
SCP
# 使用 seuratV4
.libPaths(c("/share/biosoft/R/Seurat4lib/", .libPaths()))
library(Seurat)
#library(velocyto.R)
#library(SeuratWrappers)
library(qs)
# 加载 SCP之前设置python
Sys.setenv(RETICULATE_AUTOCONFIGURE = "FALSE")
library(reticulate)
use_python("/share/biosoft/python/Python-v3.10.10/bin/python", required = TRUE)
py_config()
#跳过警告
suppressPackageStartupMessages({
#options(reticulate.conda_binary = "/path/to/conda", SCP_env_name = "SCP_env")
library(SCP)
})
#测试 官方代码
data("pancreas_sub")
print(pancreas_sub)
data("panc8_sub")
panc8_sub <- Integration_SCP(srtMerge = panc8_sub, batch = "tech", integration_method = "Seurat")
CellDimPlot(
srt = panc8_sub, group.by = c("celltype", "tech"), reduction = "SeuratUMAP2D",
title = "Seurat", theme_use = "theme_blank"
)
panc8_rename <- RenameFeatures(
srt = panc8_sub,
newnames = make.unique(capitalize(rownames(panc8_sub[["RNA"]]), force_tolower = TRUE)),
assays = "RNA"
)
srt_query <- RunKNNMap(srt_query = pancreas_sub, srt_ref = panc8_rename, ref_umap = "SeuratUMAP2D")
ProjectionPlot( #这个会报错
srt_query = srt_query, srt_ref = panc8_rename,
query_group = "SubCellType", ref_group = "celltype"
)
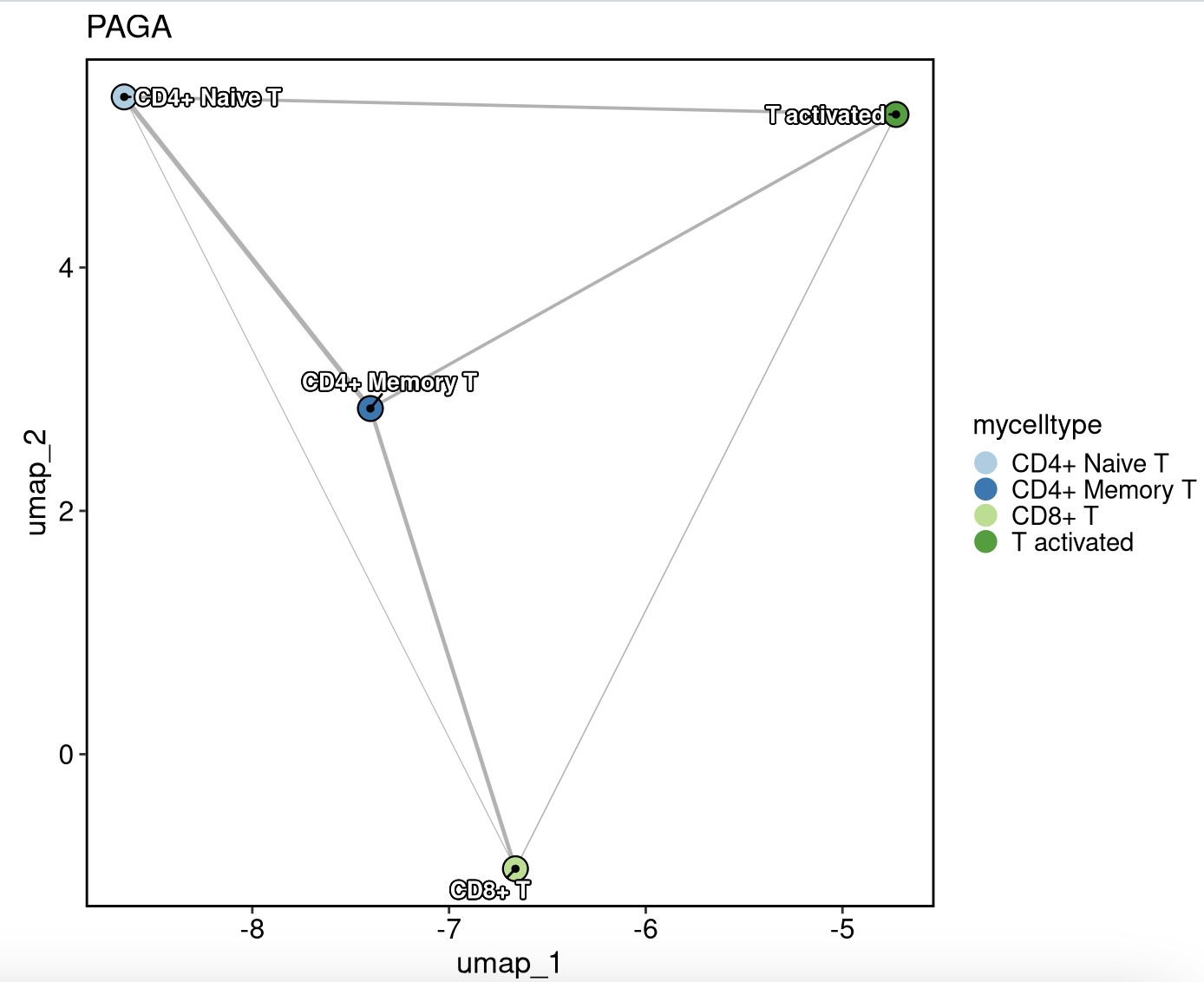
pancreas_sub <- RunPAGA(
srt = pancreas_sub, group_by = "SubCellType",
linear_reduction = "PCA", nonlinear_reduction = "UMAP"
)
PAGAPlot(srt = pancreas_sub, reduction = "UMAP", label = TRUE, label_insitu = TRUE, label_repel = TRUE)
pancreas_sub <- RunSCVELO(
srt = pancreas_sub, group_by = "SubCellType",
linear_reduction = "PCA", nonlinear_reduction = "UMAP"
)
VelocityPlot(srt = pancreas_sub, reduction = "UMAP", group_by = "SubCellType")
添加 splice 和unsplice assay
Rscript $scripts/add_usAssay_ToSeuratObj.r -i subset.T.qs -l ../01.cellranger/pbmc_ctrl/velocyto/pbmc_ctrl.loom \
../01.cellranger/pbmc_stim/velocyto/pbmc_stim.loom -p subset.T.US
R 代码
obj=qread("subset.T.US.qs")
#bm Assay5转换成 Assay
obj[["RNA4"]] <- as(object = obj[["RNA"]], Class = "Assay")
DefaultAssay(obj)<-"RNA4"
obj <- RunPAGA(
srt = obj, group_by = "mycelltype",assay_X = "RNA4",
assay_layers = c("spliced", "unspliced"),
slot_layers = "counts",
linear_reduction = "pca", nonlinear_reduction = "umap"
)
PAGAPlot(srt = obj, reduction = "UMAP", label = TRUE, label_insitu = TRUE, label_repel = TRUE)
obj <- RunSCVELO(
srt = obj, group_by = "mycelltype",assay_X = "RNA4",
linear_reduction = "pca", nonlinear_reduction = "umap"
)
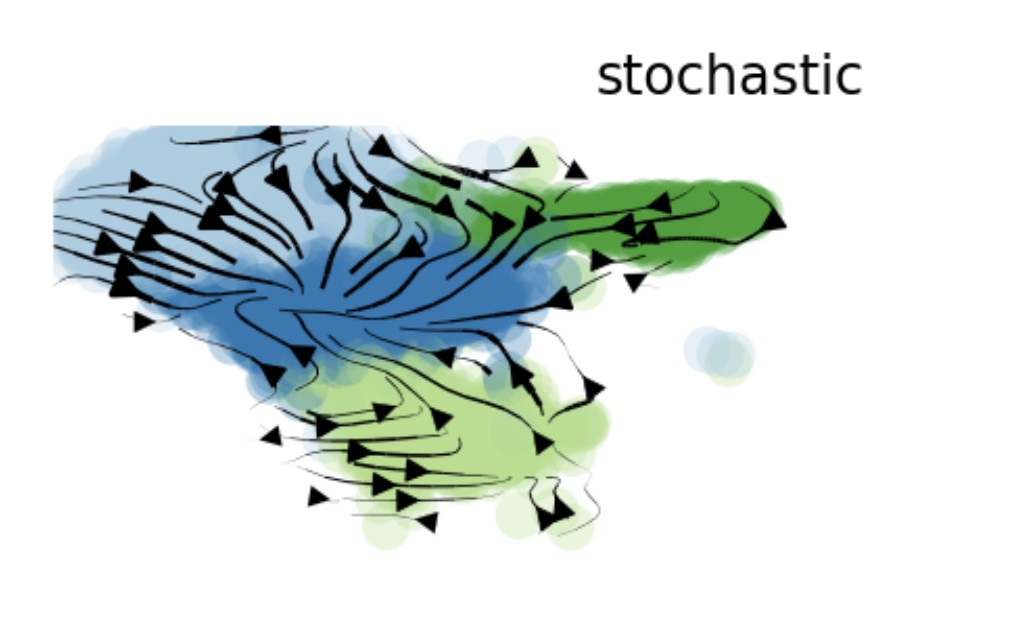
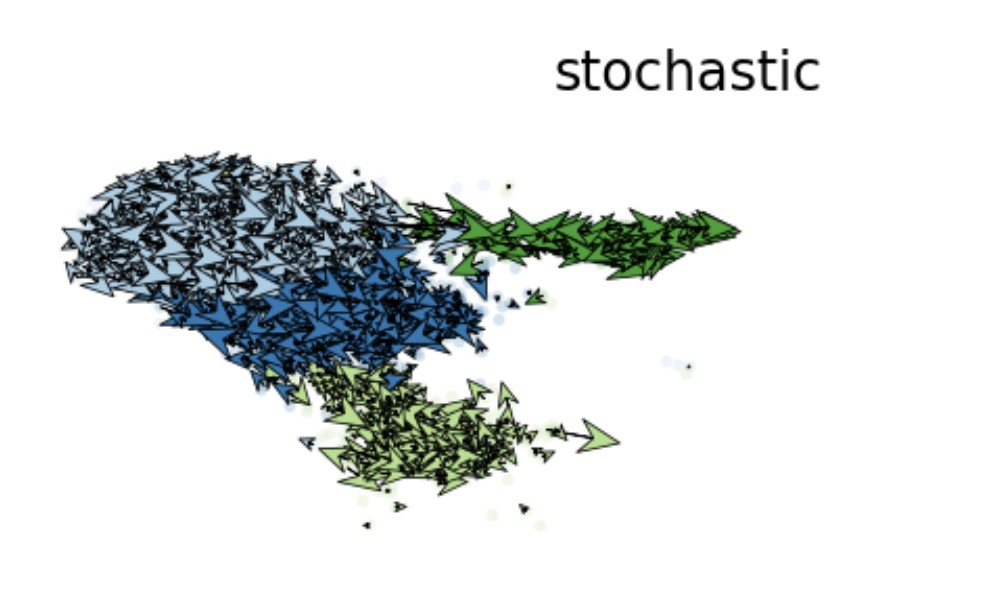
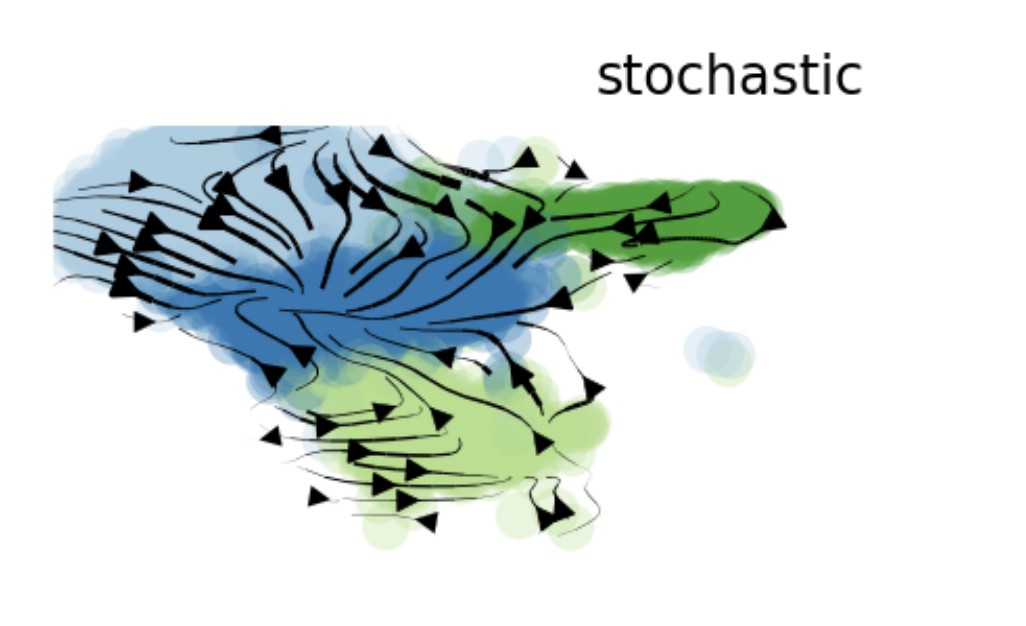
VelocityPlot(srt = obj, reduction = "umap", group_by = "mycelltype")
VelocityPlot(srt = obj, reduction = "umap", plot_type = "stream")
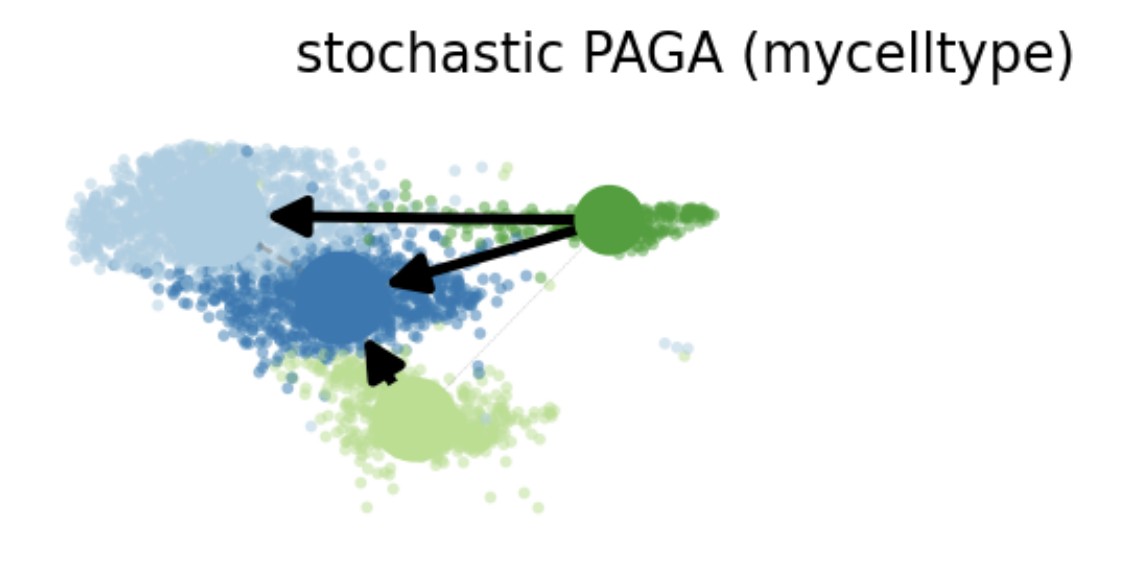
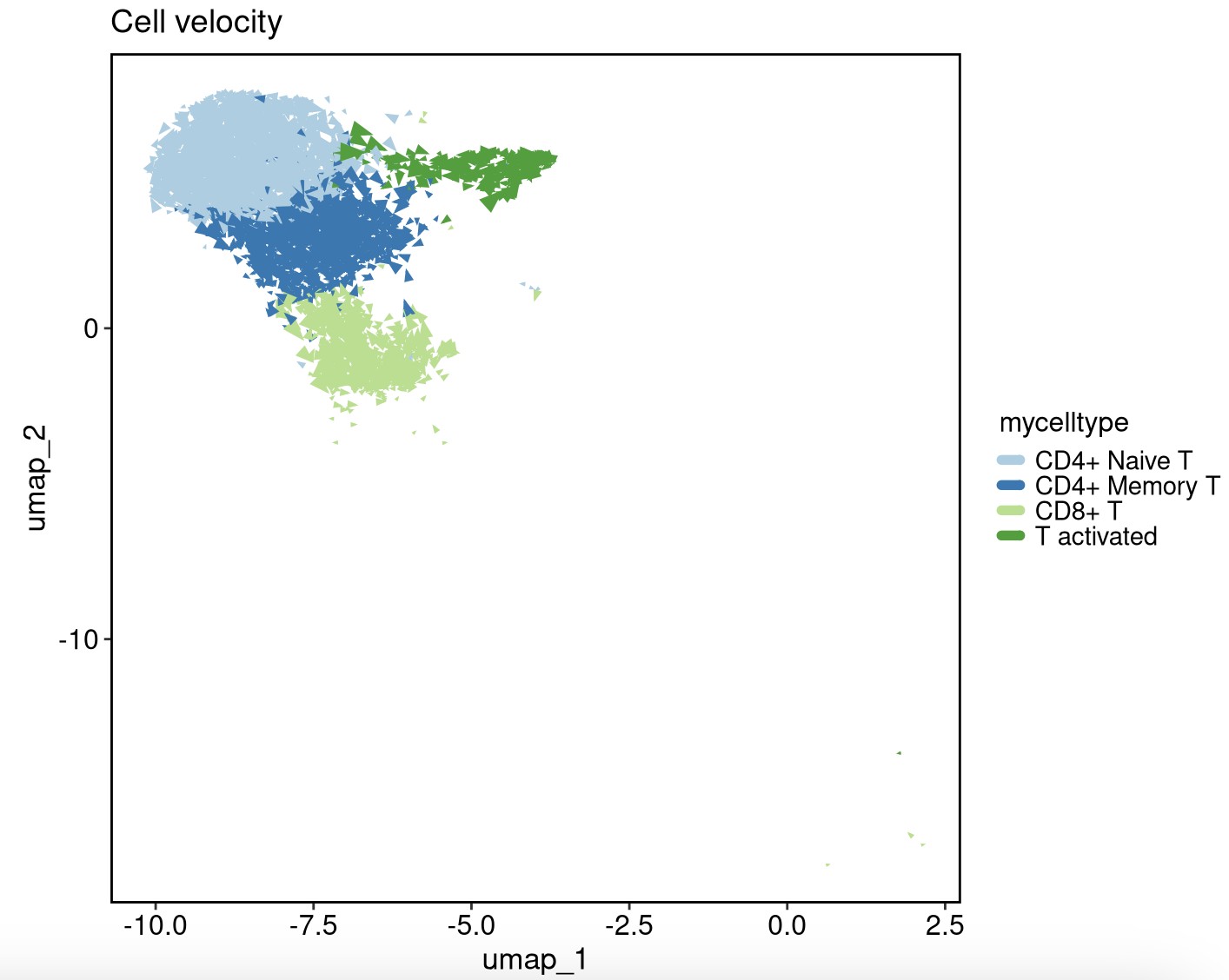
obj <- RunMonocle3(srt = obj, reduction = "umap")
names(obj@tools$Monocle3)
trajectory <- obj@tools$Monocle3$trajectory
milestones <- obj@tools$Monocle3$milestones
CellDimPlot(obj, group.by = "Monocle3_partitions", reduction = "UMAP", label = TRUE, theme_use = "theme_blank") + trajectory + milestones
CellDimPlot(obj, group.by = "Monocle3_clusters", reduction = "UMAP", label = TRUE, theme_use = "theme_blank") + trajectory
FeatureDimPlot(obj, features = "Monocle3_Pseudotime", reduction = "UMAP", theme_use = "theme_blank") + trajectory

- 发表于 2025-12-05 20:00
- 阅读 ( 385 )
- 分类:转录组
你可能感兴趣的文章
- 单细胞转录组数据标准化: 1775 浏览
- SCP FeatureDimPlot 报错 1474 浏览
- 植物单细胞转录组文章 1644 浏览
- 文件夹名称中存在空格,scp如何正确识别 1668 浏览
- 单细胞分析绘制marker基因展示图时如何设置细胞排布顺序 2020 浏览
相关问题
- 老师,图1是我用图2的代码实现的,现在我想仿照提取细胞亚类的方式,使用图3的代码单独提取TIP这个maker基因,不提取整个cluster3,这个代码可以实现吗,或者是否有其他代码可以使用 1 回答
- 老师,拟时序基础初步分析时,运行以下代码时Rscript $scripts/cell_trajectory_monocle3.r --rds ../04.cell_type_ann_leaf/leaf.added.celltype.qs \ --use.seurat.umap --groupby mycelltype -o monocle3.1 --cpu 6,出现图中报错,是因为聚类分群的时候resolution值太低了吗 2 回答
- Seurat 直接读取的表达矩阵可以是经过log2plus1和normalizaed to 10,000 counts per cell的矩阵吗?需要转化成raw count吗? 1 回答
0 条评论
请先 登录 后评论
