vt软件安装
vt安装
软件安装文档很简单:https://genome.sph.umich.edu/wiki/Vt#Installation
#this will create a directory named vt in the directory you cloned the repository 1. git clone https://github.com/atks/vt.git
#change directory to vt 2. cd vt
#run make, note that compilers need to support the c++0x standard 3. make
#you can test the build 4. make test
报错1:
gcc -g -Wall -O2 -I. -c -o cram/cram_index.o cram/cram_index.c
gcc -g -Wall -O2 -I. -c -o cram/cram_io.o cram/cram_io.c
cram/cram_io.c:61:18: error: lzma.h: No such file or directory
cram/cram_io.c: In function ‘lzma_mem_deflate’:
cram/cram_io.c:678: warning: implicit declaration of function ‘lzma_stream_buffer_bound’
cram/cram_io.c:684: error: ‘LZMA_OK’ undeclared (first use in this function)
cram/cram_io.c:684: error: (Each undeclared identifier is reported only once
cram/cram_io.c:684: error: for each function it appears in.)
cram/cram_io.c:684: warning: implicit declaration of function ‘lzma_easy_buffer_encode’
cram/cram_io.c:684: error: ‘LZMA_CHECK_CRC32’ undeclared (first use in this function)
cram/cram_io.c: In function ‘lzma_mem_inflate’:
cram/cram_io.c:694: error: ‘lzma_stream’ undeclared (first use in this function)
cram/cram_io.c:694: error: expected ‘;’ before ‘strm’
cram/cram_io.c:700: error: ‘LZMA_OK’ undeclared (first use in this function)
cram/cram_io.c:700: warning: implicit declaration of function ‘lzma_stream_decoder’
cram/cram_io.c:700: error: ‘strm’ undeclared (first use in this function)
cram/cram_io.c:700: warning: implicit declaration of function ‘lzma_easy_decoder_memusage’
cram/cram_io.c:715: warning: implicit declaration of function ‘lzma_code’
cram/cram_io.c:715: error: ‘LZMA_RUN’ undeclared (first use in this function)
cram/cram_io.c:716: error: ‘LZMA_STREAM_END’ undeclared (first use in this function)
cram/cram_io.c:728: error: ‘LZMA_FINISH’ undeclared (first use in this function)
cram/cram_io.c:737: warning: implicit declaration of function ‘lzma_end’
make[1]: *** [cram/cram_io.o] Error 1
make[1]: Leaving directory `/share/work/biosoft/vt/vt-0.57721/lib/htslib'
make: *** [lib/htslib/libhts.a] Error 1
这是因为make htslib时候找不到xz软件的include导致,找到htslib的Makefile: vi lib/htslib/Makefile 添加如下内容:
-I/share/work/biosoft/xz/xz-v5.2.3/include/
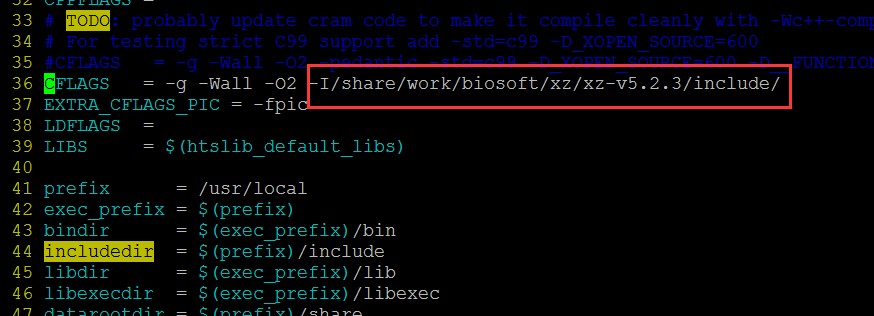
保存之后继续make。
报错2:
svm_train.cpp: In member function ‘void<unnamed>::Igor::inverse_normalize(std::vector<float, std::allocator svm_train.cpp:165: error: no matching function for call to ‘sort(__gnu_cxx::__normal_iterator<float**, std:erator<float**, std::vector<float*, std::allocator<float*> > >, <unnamed>::Igor::inverse_normalize(std::vecator<float> >&)::<anonymous struct>&)’
这是因为你的gcc版本太低,最低要求4.8.1,更换较高版本:
export PATH=/share/work/biosoft/gcc/gcc-v4.8.0/bin/:$PATH
报错3:
g++ -pipe -std=c++0x -O3 -I./lib -I. -I./lib/htslib -I./lib/Rmath -I./lib/pcre2 -I/share/work/biosoft/xz/xz-v5.2.3/include/ -D__STDC_LIMIT_MACROS -o vt ahmm.o align.o allele.o annotate_1000g.o annotate_dbsnp_rsid.o annotate_indels.o annotate_indels2.o annotate_regions.o annotate_variants.o annotate_vntrs.o augmented_bam_record.o bcf_genotyping_buffered_reader.o bcf_single_genotyping_buffered_reader.o bam_ordered_reader.o bcf_ordered_reader.o bcf_ordered_writer.o bcf_synced_reader.o bed.o candidate_motif_picker.o candidate_region_extractor.o cat.o chmm.o complex_genotyping_record.o compute_concordance.o compute_features.o compute_features2.o compute_rl_dist.o config.o consolidate_multiallelics.o consolidate_vntrs.o consolidate.o construct_probes.o decompose.o decompose2.o decompose_blocksub.o discover.o duplicate.o estimate.o estimator.o extract_vntrs.o filter.o filter_overlap.o flank_detector.o fuzzy_aligner.o fuzzy_partition.o gencode.o genome_interval.o genotype.o genotyping_record.o ghmm.o hts_utils.o hfilter.o indel_annotator.o indel_genotyping_record.o index.o info2tab.o interval_tree.o interval.o lfhmm.o lhmm.o lhmm1.o liftover.o log_tool.o merge.o merge_candidate_variants.o merge_genotypes.o milk_filter.o motif_tree.o motif_map.o multi_partition.o multiallelics_consolidator.o needle.o normalize.o nuclear_pedigree.o ordered_bcf_overlap_matcher.o ordered_region_overlap_matcher.o partition.o paste.o paste_and_compute_features_sequential.o paste_genotypes.o pedigree.o peek.o pileup.o pregex.o profile_afs.o profile_chm1.o profile_chrom.o profile_fic_hwe.o profile_hwe.o profile_indels.o profile_len.o profile_mendelian.o profile_na12878.o profile_snps.o profile_vntrs.o program.o read_filter.o reference_sequence.o rfhmm.o rfhmm_x.o rminfo.o seq.o set_ref.o snp_genotyping_record.o sort.o subset.o sv_tree.o svm_train.o svm_predict.o tbx_ordered_reader.o test.o trio.o union_variants.o uniq.o utils.o validate.o variant.o variant_manip.o variant_filter.o view.o vntr.o vntr_annotator.o vntr_consolidator.o vntr_extractor.o vntr_genotyping_record.o vntr_tree.o vntrize.o wdp_ahmm.o main.o lib/htslib/libhts.a lib/Rmath/libRmath.a lib/pcre2/libpcre2.a lib/libdeflate/libdeflate.a -lz -lpthread -lbz2 -llzma -lcurl -lcrypto
/usr/bin/ld: cannot find -llzma
collect2: error: ld returned 1 exit status
make: *** [vt] Error 1
添加library路径:
export LIBRARY_PATH=/share/work/biosoft/xz/xz-v5.2.3/lib/:$LIBRARY_PATH
再make顺利编译完成
- 发表于 2018-09-27 11:45
- 阅读 ( 5010 )
- 分类:软件工具
你可能感兴趣的文章
相关问题
0 条评论
请先 登录 后评论
