inferCNV 测试
如果你的cell来自多个病人建议按病人分组,并添加 --cluster_by_groups 参数,infer CNV会按病人分组再聚类
Rscript ../infercnv.r -i ../BCC_GSE123813.qs \
-r "T_cells" --annotations_file ../cellanno_selected.tsv --gene_location ../hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode subclusters --cluster_by_groups
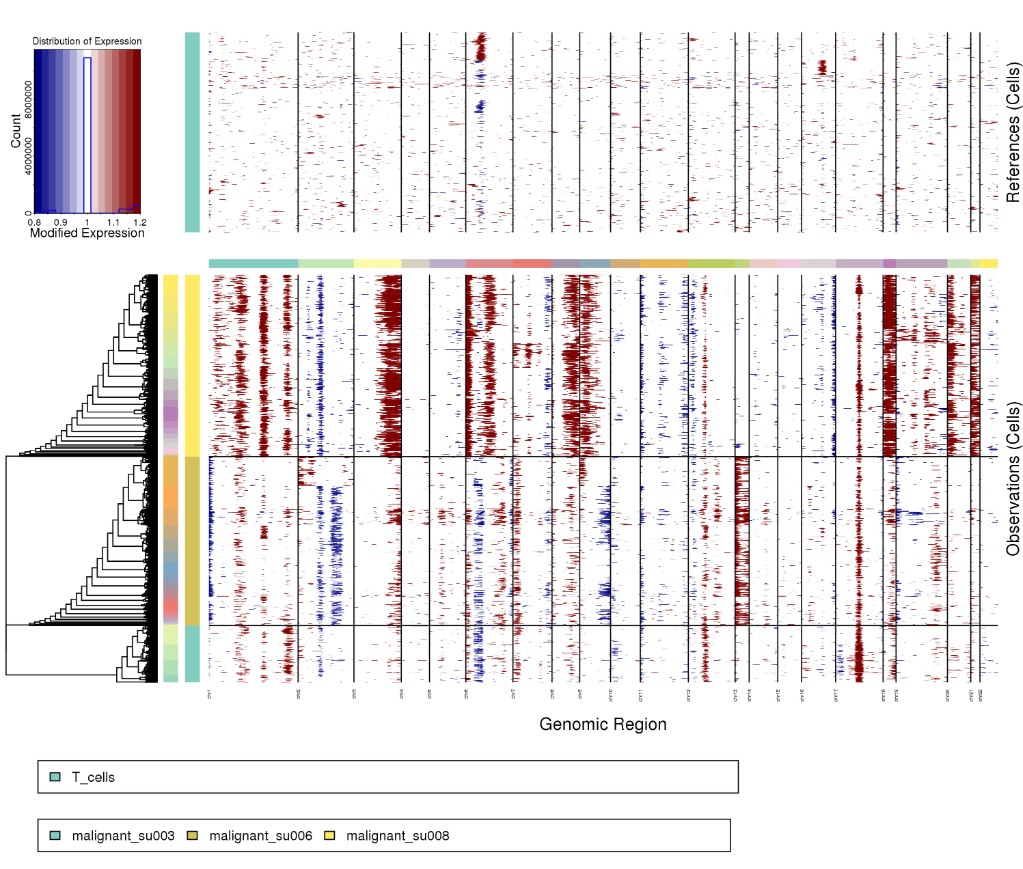
Rscript ../infercnv.r -i ../BCC_GSE123813.qs \
-r "T_cells" --annotations_file ../cellanno_selected.tsv --gene_location ../hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode subclusters
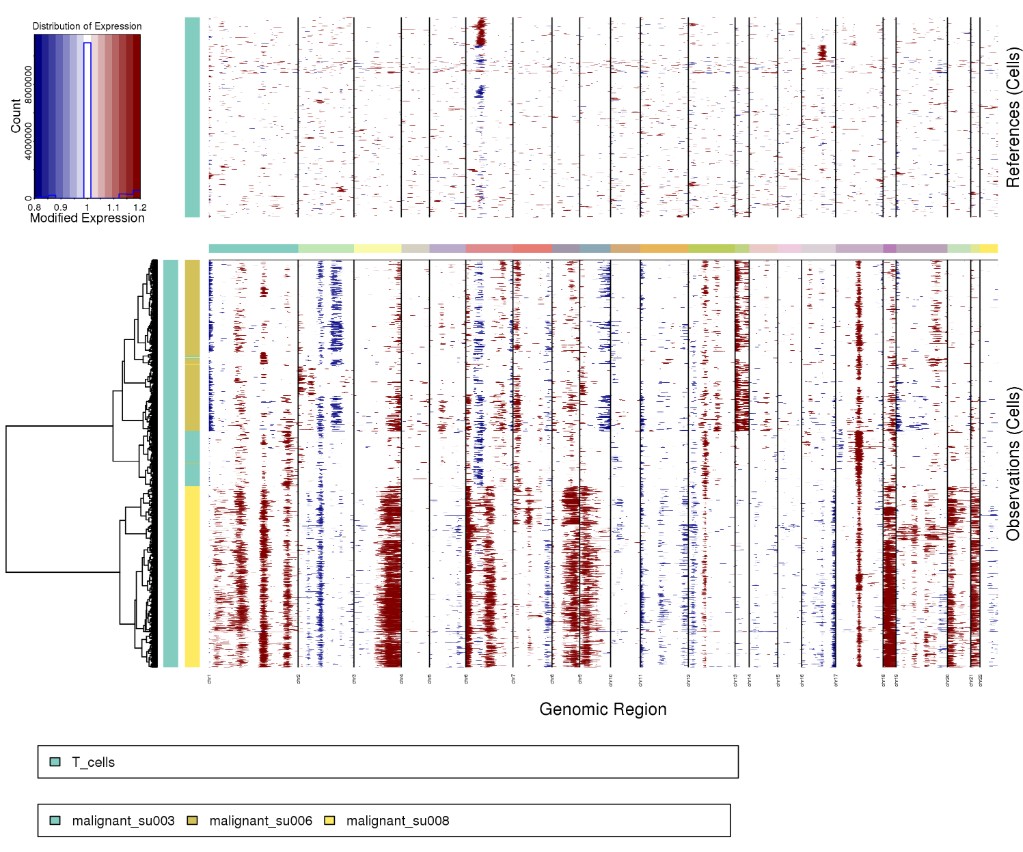
Rscript ../infercnv.r -i ../BCC_GSE123813.qs \
-r "T_cells" --annotations_file ../cellanno_selected.tsv --gene_location ../hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode samples
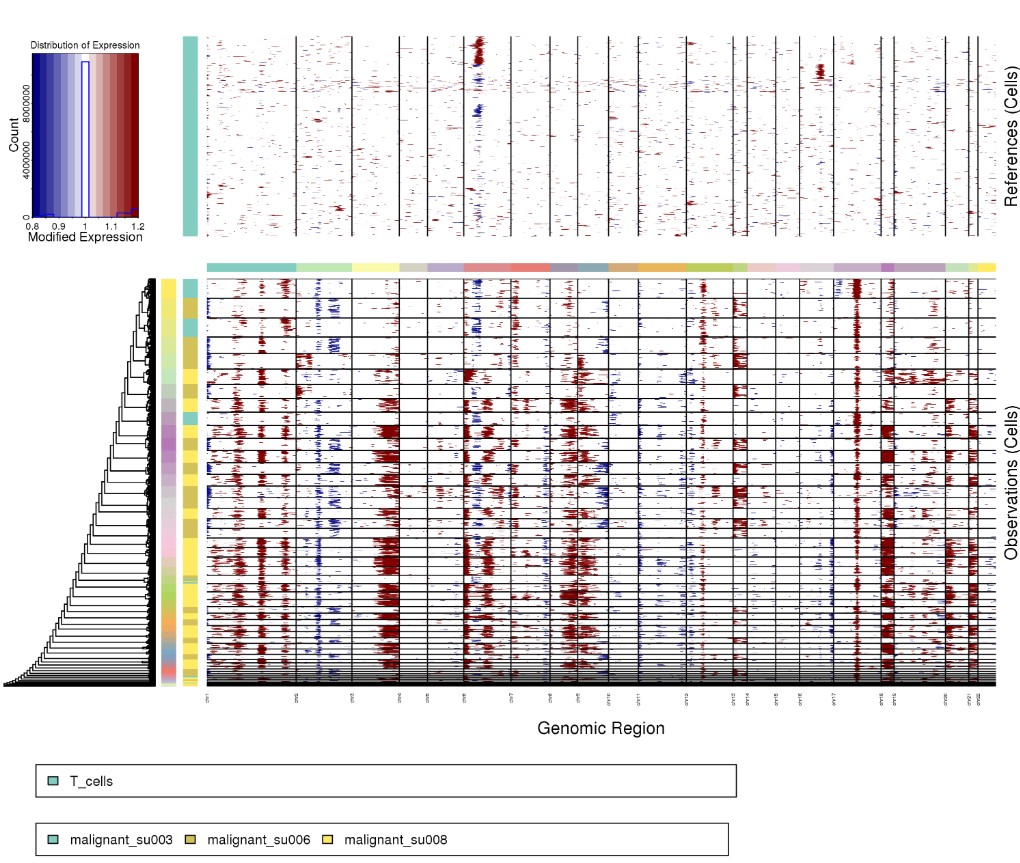
Rscript ../infercnv.r -i ../BCC_GSE123813.qs \
-r "T_cells" --annotations_file ../cellanno_selected.tsv --gene_location ../hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode samples --cluster_by_groups
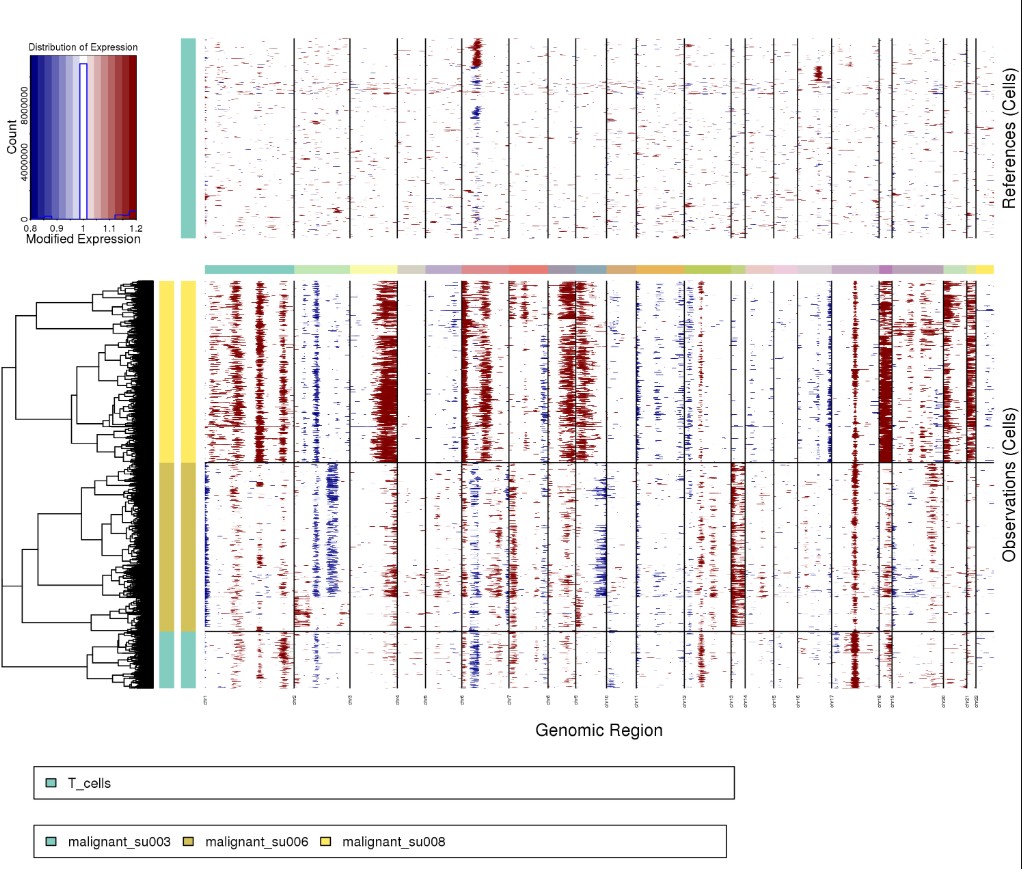
Rscript ../infercnv.r -i ../BCC_GSE123813.qs \
-r "T_cells" --annotations_file ../cellanno_selected.tsv --gene_location ../hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode subclusters --cluster_by_groups --tumor_subcluster_partition_method random_trees
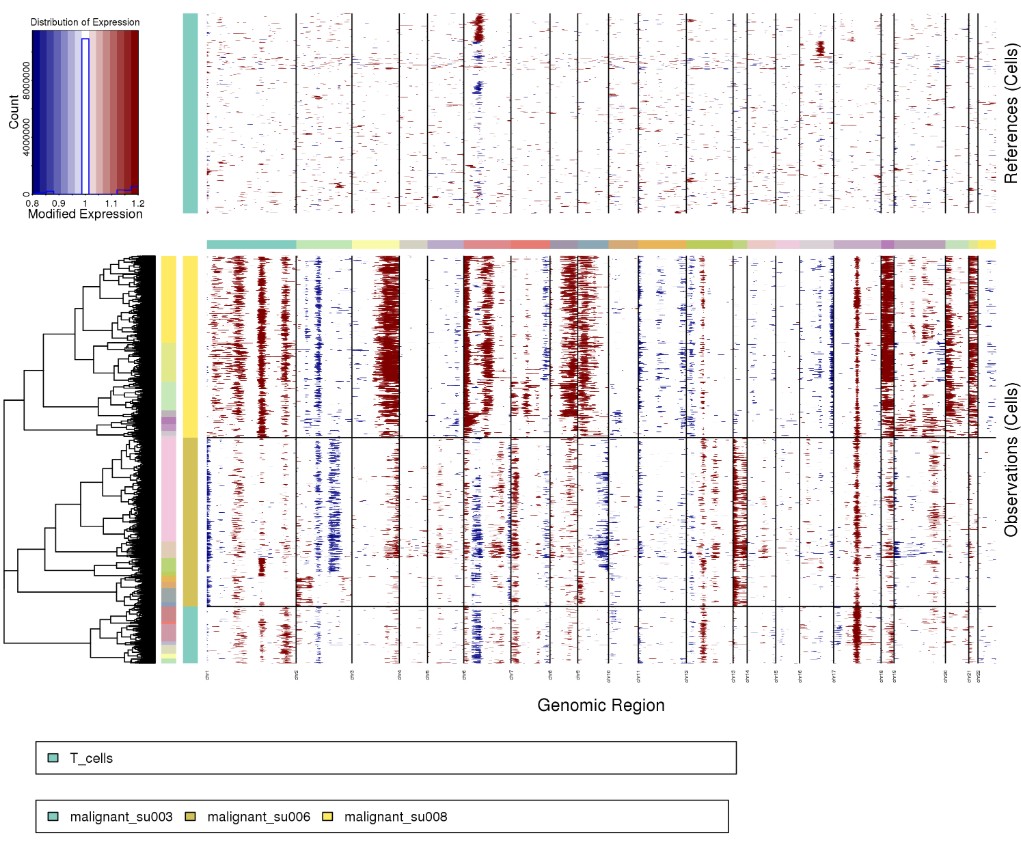
Rscript ../infercnv.r -i ../BCC_GSE123813.qs \
-r "T_cells" --annotations_file ../cellanno_selected.tsv --gene_location ../hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode subclusters --tumor_subcluster_partition_method random_trees
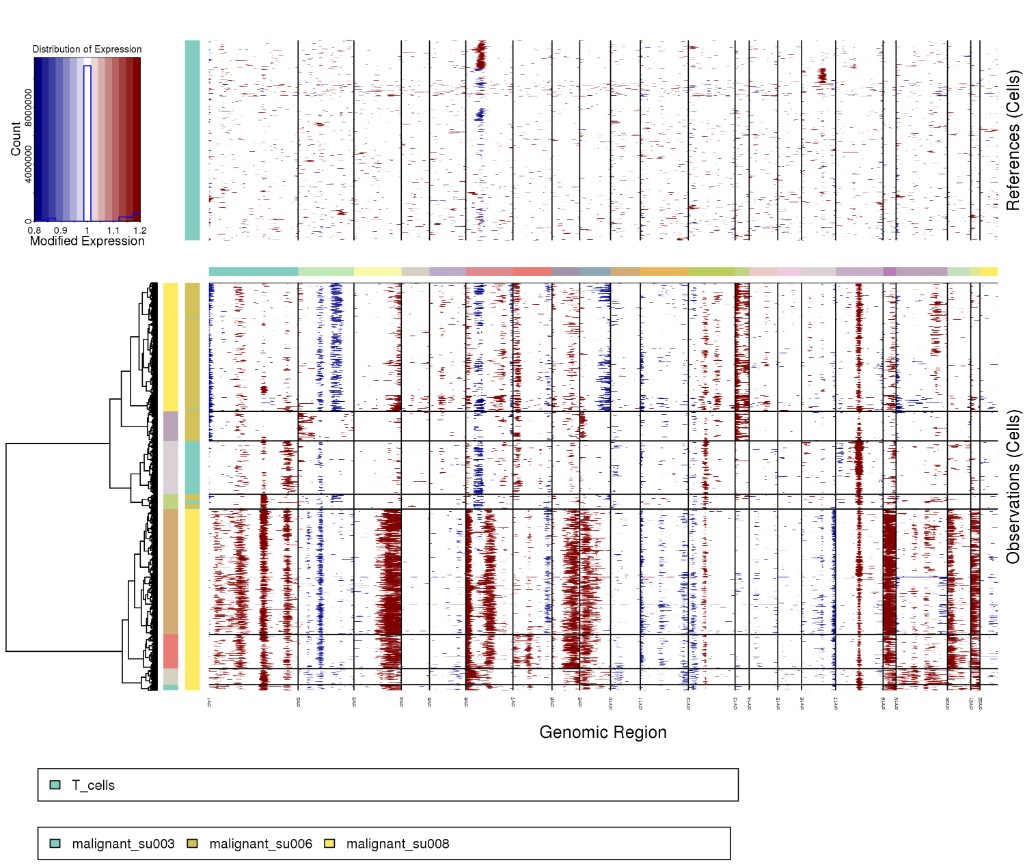
Rscript ../infercnv.r -i ../BCC_GSE123813.qs \
-r "T_cells" --annotations_file ../cellanno_selected.tsv --gene_location ../hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode samples --cluster_by_groups --tumor_subcluster_partition_method random_trees
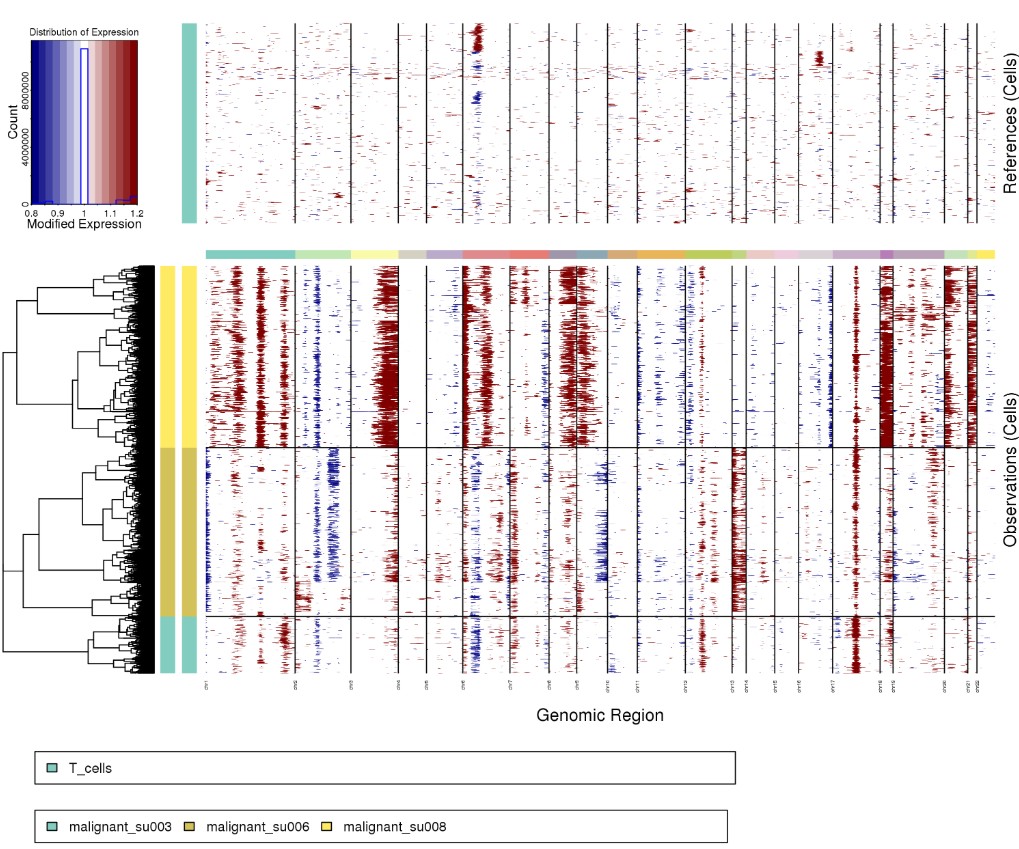
Rscript ../infercnv.r -i ../BCC_GSE123813.qs \
-r "T_cells" --annotations_file ../cellanno_selected.tsv --gene_location ../hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode samples --tumor_subcluster_partition_method random_trees
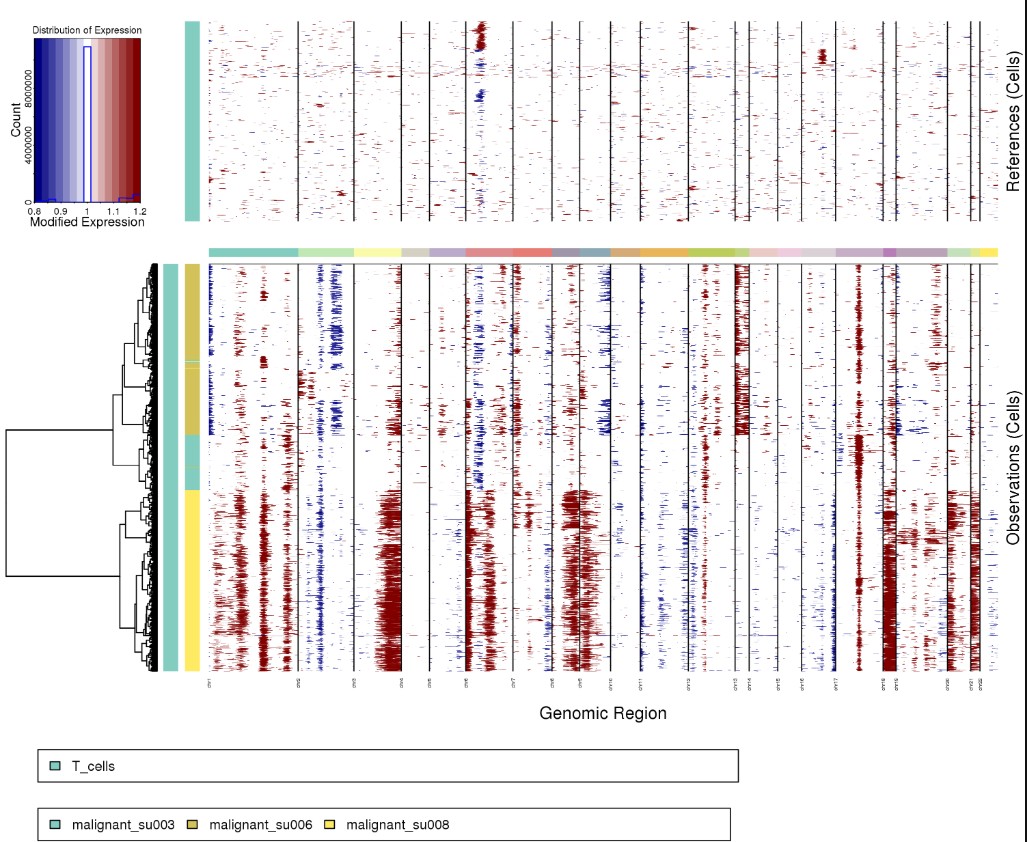
Rscript infercnv.r -i ../12.niche/molecular_niche.GraphST_clusters.qs \
-r "normal" --annotations_file cellanno.tsv --gene_location hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode subclusters --k_obs_groups 5 \
--hmmstate i6 -o infercnv3
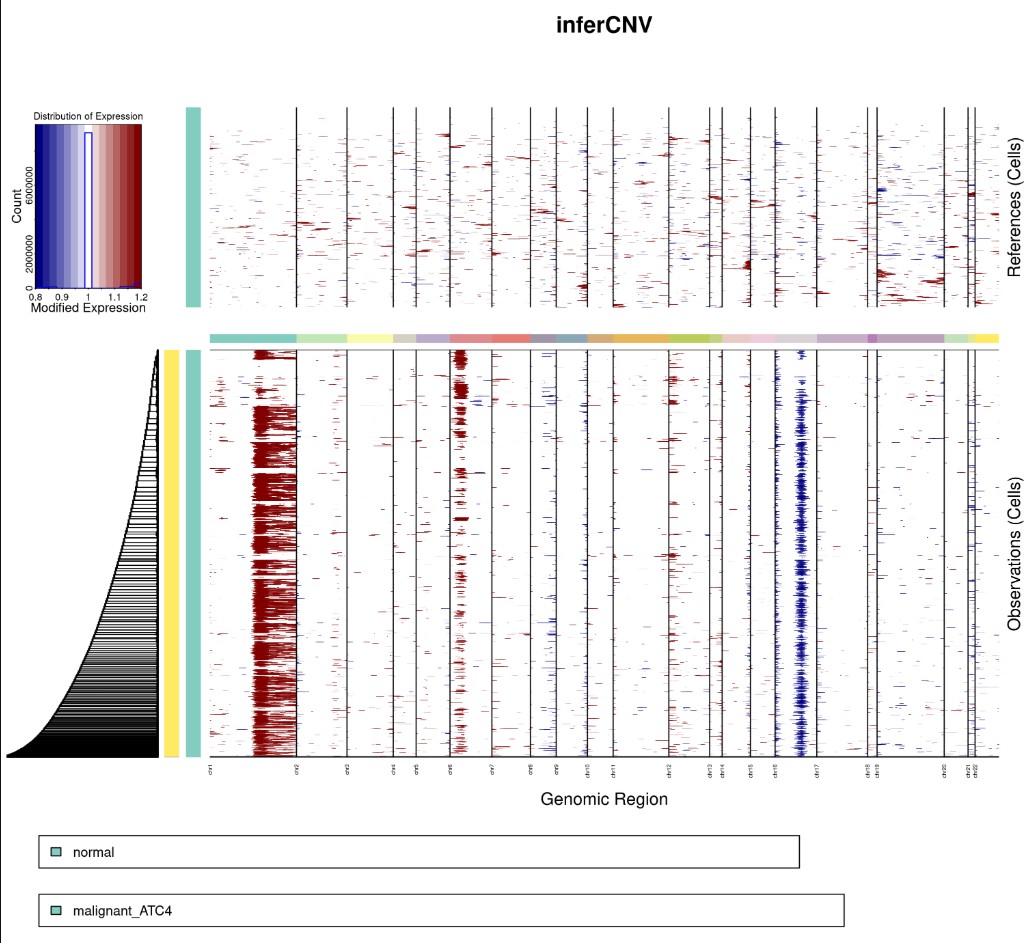
Rscript infercnv.r -i ../12.niche/molecular_niche.GraphST_clusters.qs \
-r "normal" --annotations_file cellanno.tsv --gene_location hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode samples --k_obs_groups 5 \
--hmmstate i6 -o infercnv4
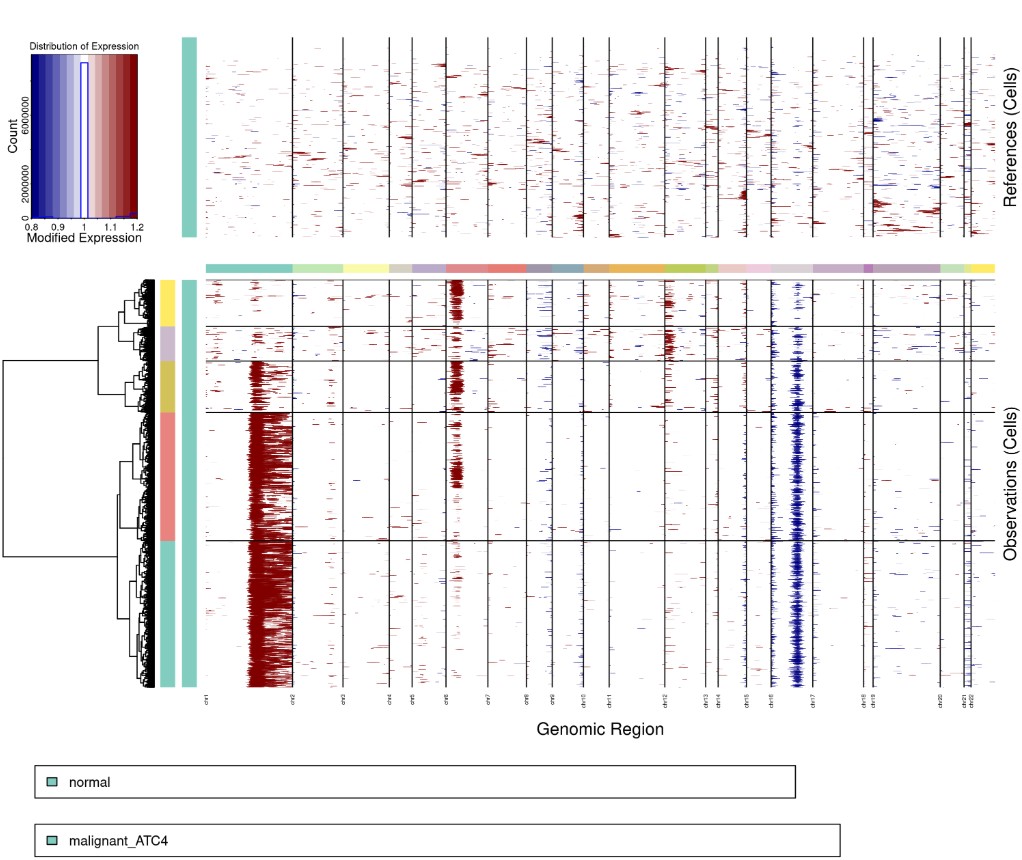
Rscript infercnv.r -i ../12.niche/molecular_niche.GraphST_clusters.qs \
-r "normal" --annotations_file cellanno.tsv --gene_location hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode samples --k_obs_groups 5 --tumor_subcluster_partition_method leiden \
--hmmstate i6 -o infercnv5

Rscript infercnv.r -i ../12.niche/molecular_niche.GraphST_clusters.qs \
-r "normal" --annotations_file cellanno.tsv --gene_location hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode subclusters --tumor_subcluster_partition_method leiden --leiden_resolution 0.01 \
--hmmstate i6 -o infercnv7
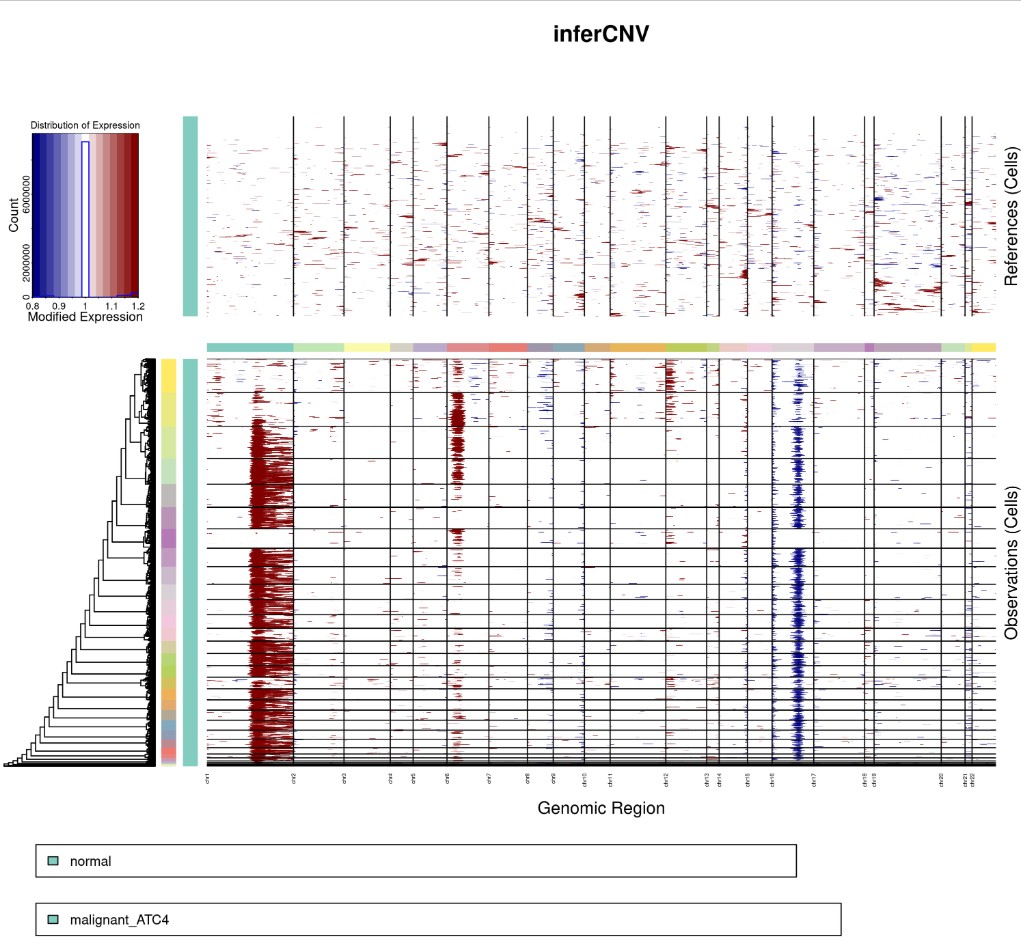
Rscript infercnv.r -i ../12.niche/molecular_niche.GraphST_clusters.qs \
-r "normal" --annotations_file cellanno.tsv --gene_location hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode samples --tumor_subcluster_partition_method leiden --leiden_resolution 0.01 \
--hmmstate i6 -o infercnv6
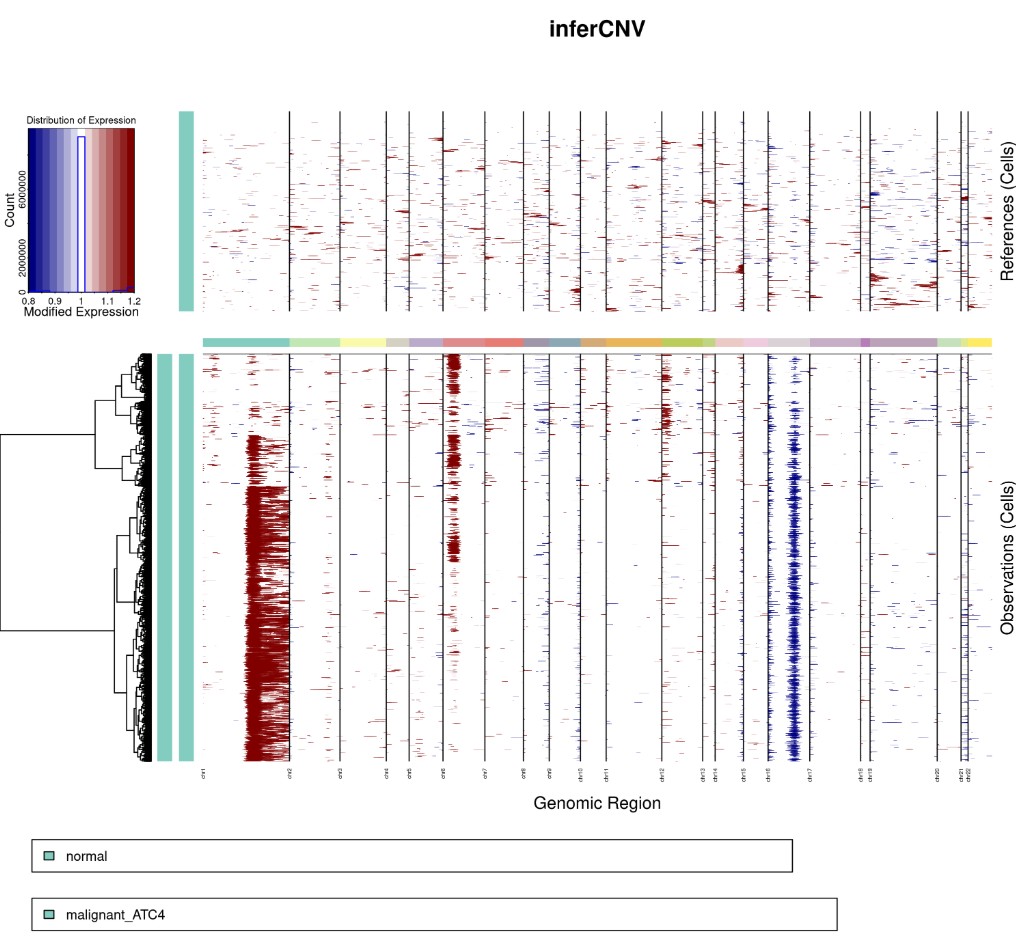
## leiden 聚类方法 --leiden_resolution 0.01 控制聚类数量 通常很多
Rscript infercnv.r -i ../12.niche/molecular_niche.GraphST_clusters.qs \
-r "normal" --annotations_file cellanno.tsv --gene_location hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode subclusters --cluster_by_groups \
--tumor_subcluster_partition_method leiden --leiden_resolution 0.001 \
--hmmstate i6 -o infercnv3
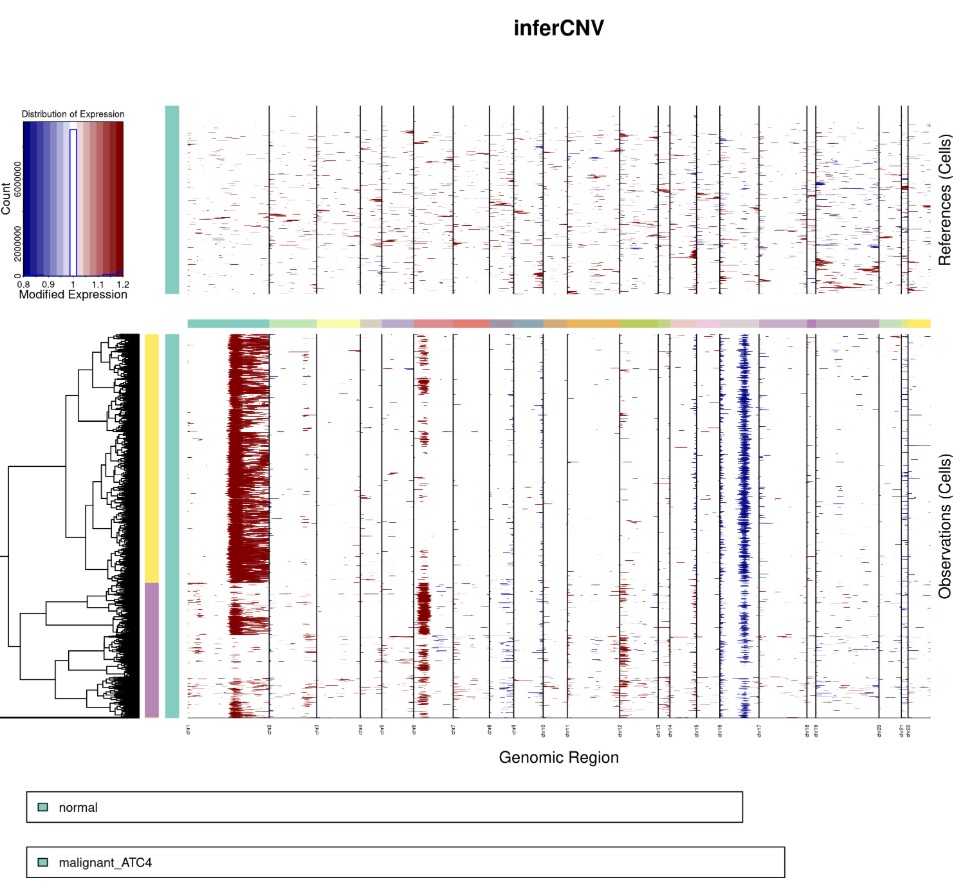
# random_trees 聚类方法最多 8类 肿瘤克隆进化分析需要
Rscript infercnv.r -i ../12.niche/molecular_niche.GraphST_clusters.qs \
-r "normal" --annotations_file cellanno.tsv --gene_location hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode subclusters --cluster_by_groups \
--tumor_subcluster_partition_method random_trees --hmmstate i6 -o infercnv2
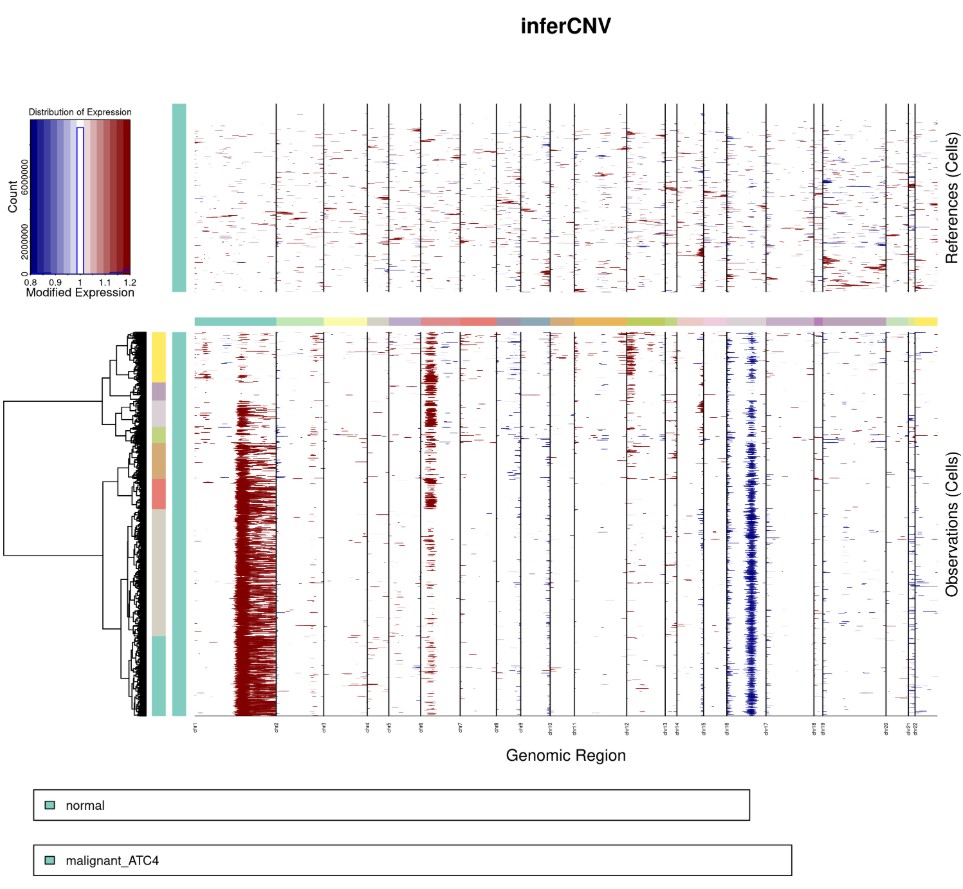
Rscript infercnv.r -i ../12.niche/molecular_niche.GraphST_clusters.qs \
-r "normal" --annotations_file cellanno.tsv --gene_location hg38_gencode_v27.txt \
--cpu 20 --hmm --denoise --analysis_mode subclusters --cluster_by_groups --k_obs_groups 5 \
--hmmstate i6 -o infercnv1
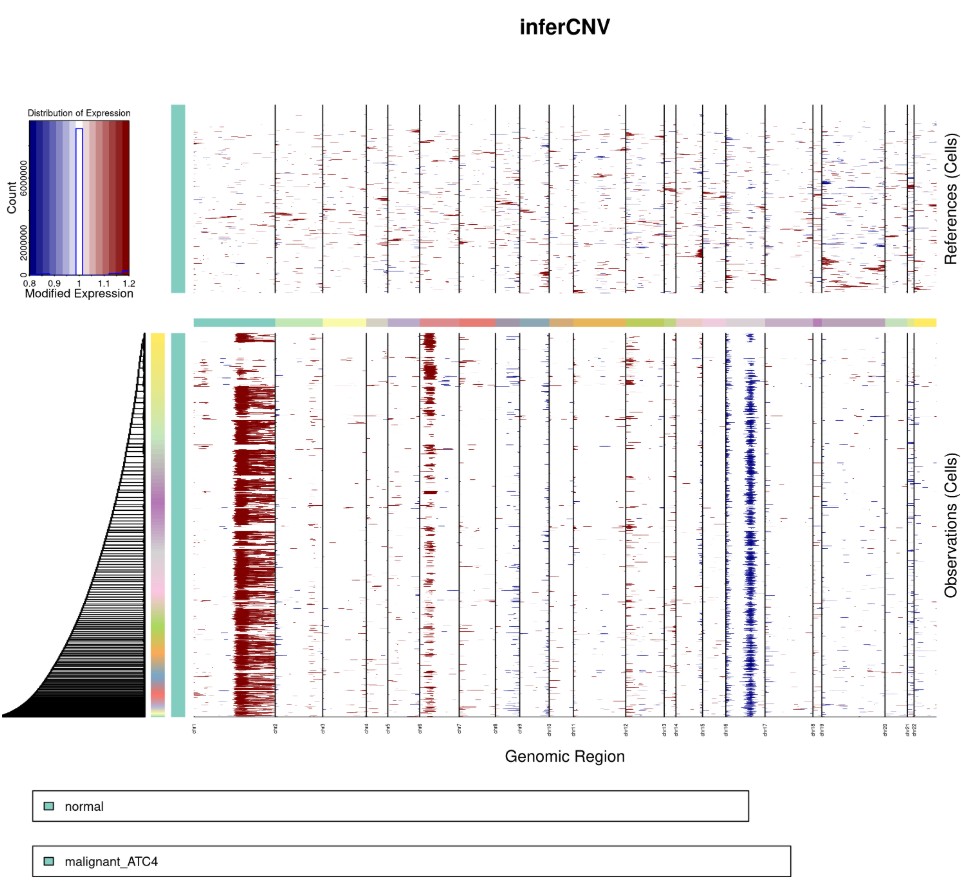

- 发表于 2025-01-10 11:26
- 阅读 ( 1635 )
- 分类:转录组
