从gff文件当中提取对应转录本ID的基因结构信息用于GSDS绘制结构图脚本更新
get_gene_exon_from_gff.pl 脚本更新,
脚本名称:get_gene_exon_from_gff.pl
用法:perl ../script/get_gene_exon_from_gff.pl -in1 WRKY_domain_new_out_emoved_redundant.txt -in2 Arabidopsis_thaliana.TAIR10.31.gff3 -out gene_exon_info.gff
输出结果(部分):
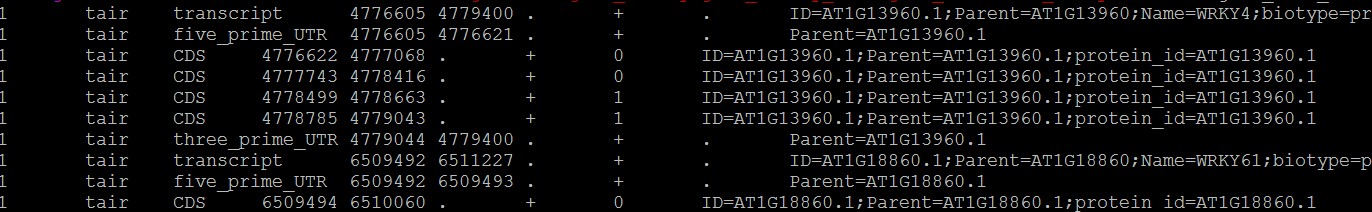
脚本源代码:
use Getopt::Long;
my %opts;
use Data::Dumper;
GetOptions( \%opts, "in1=s", "in2=s", "out=s", "h" );
if ( !defined( $opts{in1} )
|| !defined( $opts{in2} )
|| !defined( $opts{out} )
|| defined( $opts{h} ) )
{
&USAGE;
}
open( IN1, "$opts{in1}" ) || die "open $opts{in1} failed\n";
open( IN2, "$opts{in2}" ) || die "open $opts{in2} failed\n";
open( OUT, ">$opts{out}" ) || die "open $opts{out} failed\n";
my %gffs;
while (<IN1>) {
chomp;
next if /^#/;
my @b = split/\t/, $_;
$gffs{$b[0]} = 1;
}
#print Dumper(\%gffs);
while (<IN2>) {
chomp;
next if (/^#/);
my @a = split /\t/, $_;
next if $a[2]=~/exon/i;
if ($a[2] =~/^mRNA$/i or $a[2] =~/^transcript$/i ) {
($id1) = ($a[8] =~ m/ID=([^;]*)/);
}elsif ( $a[2] =~/^CDS$/i or $a[2] =~/utr/i ) {
($id1) = ($a[8] =~ m/Parent=([^;]*)/);
}else{
next;
}
if ( exists $gffs{$id1} ) {
print OUT "$_\n";
}
}
close OUT;
close IN1;
close IN2;
sub USAGE {
print "usage: perl $0 -in1 mRNA_id.txt -in2 genome.gff3 -out gene_location.txt ";
exit;
}
脚本下载地址:get_gene_exon_from_gff.pl 可下载替换以前的版本。
- 发表于 2019-03-15 13:34
- 阅读 ( 8273 )
- 分类:perl
你可能感兴趣的文章
- IF=4.8|对4种天南星科植物基因组进行WRKY基因家族分析,并分析其在魔芋中的表达 274 浏览
- IF=4.3 | 秋茄树SOS1基因家族分析 2154 浏览
- IF=4.9 | 多倍体割手密IAA基因家族分析鉴定 2579 浏览
- GFF或GTF格式转bed 2883 浏览
- 紧扣前沿热点--多种研究培训,总有你需要的! 6075 浏览
- 2024最新版《基因家族分析实操》直播培训开始报名了! 3587 浏览
1 条评论
请先 登录 后评论
