KEGG GO 富集显著性图 名字太长,柱状图上文字解决
KEGG GO 富集显著性图 名字太长,柱状图上文字解决
library(ggplot2)
library(dplyr)
library(forcats)
# 创建数据框
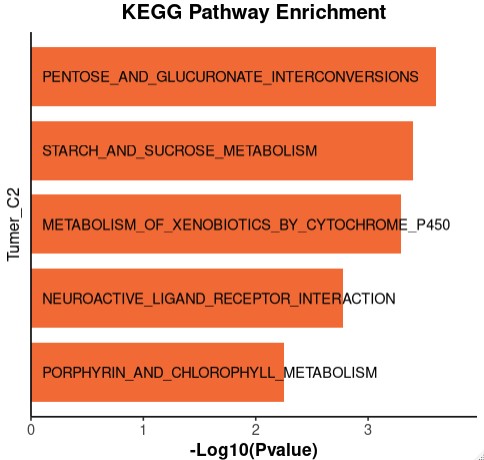
data <- data.frame(
Description = c(
"KEGG_STARCH_AND_SUCROSE_METABOLISM",
"KEGG_PENTOSE_AND_GLUCURONATE_INTERCONVERSIONS",
"KEGG_METABOLISM_OF_XENOBIOTICS_BY_CYTOCHROME_P450",
"KEGG_PORPHYRIN_AND_CHLOROPHYLL_METABOLISM",
"KEGG_NEUROACTIVE_LIGAND_RECEPTOR_INTERACTION"
),
pvalue = c(0.000396688, 0.000246396, 0.000508482, 0.005582234, 0.001678246)
)
# 数据处理:计算-log10(pvalue)并排序
data <- data %>%
mutate(
Description_short = gsub("KEGG_", "", Description),
log_pvalue = -log10(pvalue)
) %>%
arrange(log_pvalue) %>% # 按升序排列,这样最大的在顶部
mutate(Description_short = fct_reorder(Description_short, log_pvalue))
# 创建条形图
ggplot(data, aes(x = log_pvalue, y = Description_short)) +
geom_bar(stat = "identity", fill = "#F0510C", width = 0.8,alpha=0.8) +
geom_text(aes(label = Description_short, x = 0.1),
hjust = 0, size = 3.5, color = "black") +
labs(
title = "KEGG Pathway Enrichment",
x = "-Log10(Pvalue)",
y = "Tumer_C2"
) +
theme_classic() +
theme(
plot.title = element_text(size = 14, face = "bold", hjust = 0.5),
axis.title.x = element_text(size = 12, face = "bold"),
axis.text.y = element_blank(), # 隐藏Y轴文字
axis.ticks.y = element_blank(), # 隐藏Y轴刻度
axis.text.x = element_text(size = 10),
panel.grid.major.y = element_blank(),
panel.grid.minor.y = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank()
) +
scale_x_continuous(expand = expansion(mult = c(0, 0.1)))
- 发表于 2025-09-24 11:35
- 阅读 ( 1615 )
- 分类:转录组
你可能感兴趣的文章
- KEGG 通路基因列表; 1439 浏览
- metascape数据库 8890 浏览
- eggNOG批量注释基因组数据库 5771 浏览
- clusterProfiler非模式物种 KEGG与GO富集分析 16206 浏览
- enrichGSEA_pip.r GSEA富集分析 4075 浏览
0 条评论
请先 登录 后评论
