您好,“或者直接用新的脚本覆盖掉之前的,就能解决”,请问新的脚本是什么呢?
ANNOVAR变异注释报错:Erro: invalid record found in exonic_variant_function file (exonic format error)
老师您好,
我的输入代码: table_annovar.pl $workdir/4.snp_indel/var_qc/all.clean.snp.vcf.gz $refdir/test \ -buildver unknown -out $workdir/5.var_ann/test/snp \ -remove -protocol refGene -operation g -nastring . -vcfinput
报错所有信息:NOTICE: the --polish argument is set ON automatically (use --nopolish to change this behavior)
NOTICE: Running with system command <convert2annovar.pl -includeinfo -allsample -withfreq -format vcf4 /work/bdorsalis/4.snp_indel/var_qc/all.clean.snp.vcf.gz > /work/bdorsalis/5.var_ann/test/snp.avinput>
NOTICE: Finished reading 840 lines from VCF file
NOTICE: A total of 224 locus in VCF file passed QC threshold, representing 224 SNPs (152 transitions and 72 transversions) and 0 indels/substitutions
NOTICE: Finished writing allele frequencies based on 1344 SNP genotypes (912 transitions and 432 transversions) and 0 indels/substitutions for 6 samples
NOTICE: Running with system command </share/work/biosoft/annovar/latest/table_annovar.pl /work/bdorsalis/5.var_ann/test/snp.avinput /work/bdorsalis/ref/test -buildver unknown -outfile /work/bdorsalis/5.var_ann/test/snp -remove -protocol refGene -operation g -nastring . -otherinfo>
NOTICE: the --polish argument is set ON automatically (use --nopolish to change this behavior)
-----------------------------------------------------------------
NOTICE: Processing operation=g protocol=refGene
NOTICE: Running with system command <annotate_variation.pl -geneanno --neargene 1000 -buildver unknown -dbtype refGene -outfile /work/bdorsalis/5.var_ann/test/snp.refGene -exonsort -nofirstcodondel /work/bdorsalis/5.var_ann/test/snp.avinput /work/bdorsalis/ref/test>
NOTICE: Output files are written to /work/bdorsalis/5.var_ann/test/snp.refGene.variant_function, /work/bdorsalis/5.var_ann/test/snp.refGene.exonic_variant_function
NOTICE: Reading gene annotation from /work/bdorsalis/ref/test/unknown_refGene.txt ... Done with 34330 transcripts (including 4228 without coding sequence annotation) for 16570 unique genes
NOTICE: Processing next batch with 224 unique variants in 224 input lines
NOTICE: Reading FASTA sequences from /work/bdorsalis/ref/test/unknown_refGeneMrna.fa ... Done with 77 sequences
WARNING: A total of 151 sequences will be ignored due to lack of correct ORF annotation
NOTICE: Running with system command <coding_change.pl /work/bdorsalis/5.var_ann/test/snp.refGene.exonic_variant_function.orig /work/bdorsalis/ref/test/unknown_refGene.txt /work/bdorsalis/ref/test/unknown_refGeneMrna.fa -alltranscript -out /work/bdorsalis/5.var_ann/test/snp.refGene.fa -newevf /work/bdorsalis/5.var_ann/test/snp.refGene.exonic_variant_function>
Error: invalid record found in exonic_variant_function file (exonic format error): <line3 synonymous SNV gene-LOC105223489:rna-XM_049455802.1#NC_064303.1#9685699:exon4:c.T1656C:p.F552F,gene-LOC105223489:rna-XM_049455807.1#NC_064303.1#9685699:exon5:c.T1656C:p.F552F NC_064303.1 9687053 9687053 A G 0.8 404.75 5 NC_064303.1 9687053 . A G 404.75 PASS AC=8;AF=0.800;AN=10;BaseQRankSum=-0.581;DP=19;ExcessHet=3.5218;FS=0.000;MLEAC=8;MLEAF=0.800;MQ=60.00;MQRankSum=0.000;QD=21.30;ReadPosRankSum=-0.185;SOR=0.368 GT:AD:DP:GQ:PL 1/1:0,3:3:9:59,9,0 1/1:0,4:4:12:133,12,0 ./.:.:.:.:. 0/1:1,2:3:33:40,0,33 0/1:2,2:4:37:37,0,37 1/1:0,5:5:15:141,15,0> at /share/work/biosoft/annovar/latest/coding_change.pl line 77, <EVF> line 1.
Error running system command: <coding_change.pl /work/bdorsalis/5.var_ann/test/snp.refGene.exonic_variant_function.orig /work/bdorsalis/ref/test/unknown_refGene.txt /work/bdorsalis/ref/test/unknown_refGeneMrna.fa -alltranscript -out /work/bdorsalis/5.var_ann/test/snp.refGene.fa -newevf /work/bdorsalis/5.var_ann/test/snp.refGene.exonic_variant_function>
Error running system command: </share/work/biosoft/annovar/latest/table_annovar.pl /work/bdorsalis/5.var_ann/test/snp.avinput /work/bdorsalis/ref/test -buildver unknown -outfile /work/bdorsalis/5.var_ann/test/snp -remove -protocol refGene -operation g -nastring . -otherinfo>
以及在前边用sh index.sh生成unknown_refGeneMrna.fa和unknown_refGene.txt也有报错:no exons defined for group , feature gene,
当时按照提示解决的:-ignoreGroupsWithoutExons
不知道是不是这里的问题影响了后边?
这个问题我在Github上查了一下,有个人提问作者,和我的问题是一样的
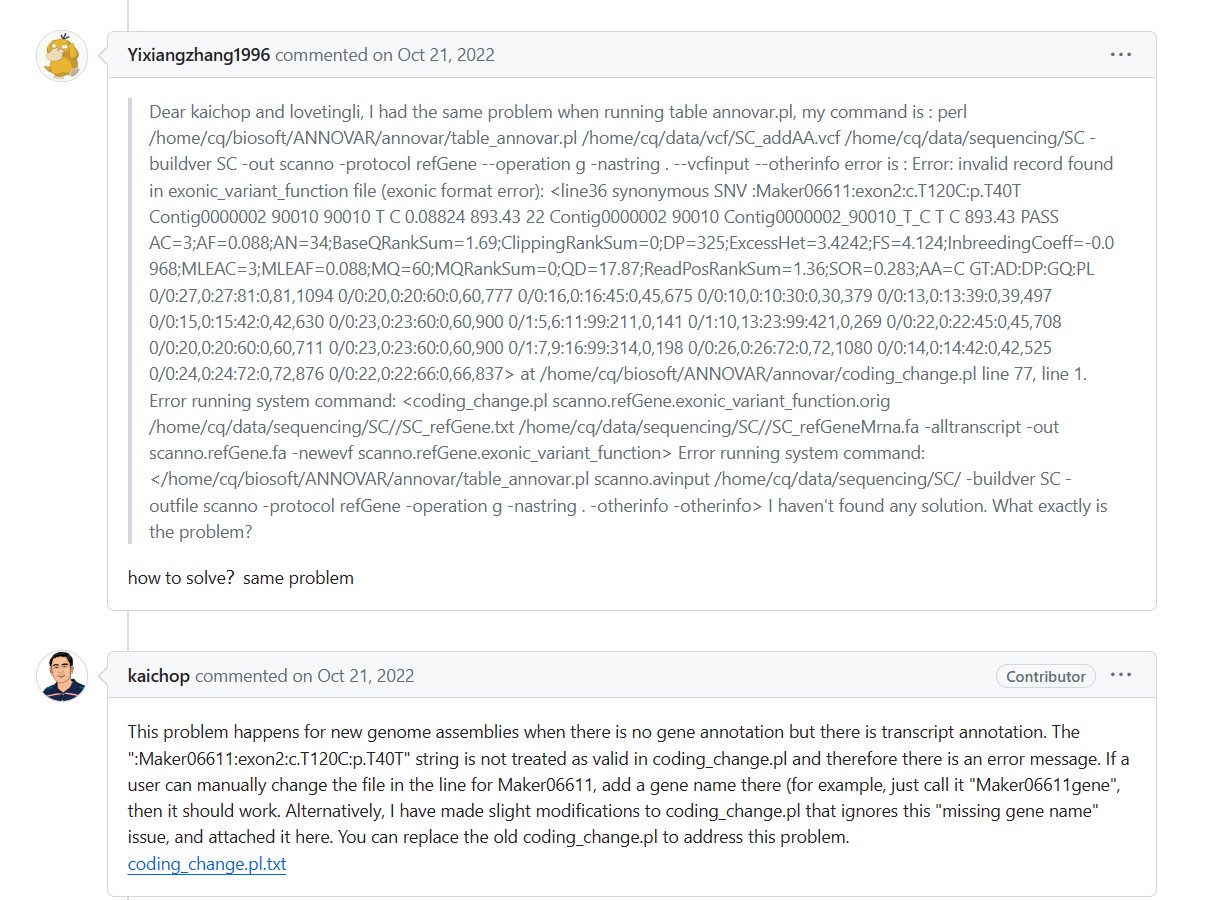 作者给出的回答是手动改掉有问题的那三行,给后边加个基因名字?不知道是改哪个文件,我在snp.refGene.variant_function中没有找到有问题的那一行(synonymous SNV gene-LOC105223489),或者直接用新的脚本覆盖掉之前的,就能解决,我尝试了 ,由于租的组学大讲堂的云服务器,没有权限,不知道我们所使用的docker镜像和云服务器中的脚本是否是最新的?
作者给出的回答是手动改掉有问题的那三行,给后边加个基因名字?不知道是改哪个文件,我在snp.refGene.variant_function中没有找到有问题的那一行(synonymous SNV gene-LOC105223489),或者直接用新的脚本覆盖掉之前的,就能解决,我尝试了 ,由于租的组学大讲堂的云服务器,没有权限,不知道我们所使用的docker镜像和云服务器中的脚本是否是最新的?看到也有别的人提了相似的问题,可能在refGene,cytoBand,1000g2014oct_eur,1000g2014oct_afr,exac03,ljb26_all,clinvar_20140929,snp138中找不到的参考的物种蛮多的?(参考基因组是在NCBI下载的)不知道老师们能不能出一期视频讲一下如何解决这个问题,以及怎么手动修改错误的数据
请先 登录 后评论
- 1 关注
- 0 收藏,2989 浏览
- 薄信 提出于 2023-12-22 19:38