输入的文件有问题吧,你对比下例子的文件
600 进行bugbase运行时候老报错
回答问题即可获得 10 经验值,回答被采纳后即可获得 605 金币。
run.bugbase.r -i pick_closed_reference_otus/table.from_txt_json.biom -m pick_closed_reference_otus/fastmap.txt -c loc -o bugbase_loc
--> Q&A for bioinformatics, please visit the website: www.omicsclass.com
--> Recommended to learn R language:
--> https://study.163.com/course/introduction/1209073807.htm?share=1&shareId=1030291076
[1] "Loading Inputs..."
[1] "16S copy number normalizing OTU table..."
[1] "Predicting phenotypes..."
Error in single.cell.predictions(trait_table, normalized_otus, test_trait, :
Error: no OTU overlap between OTU table and trait table.
Execution halted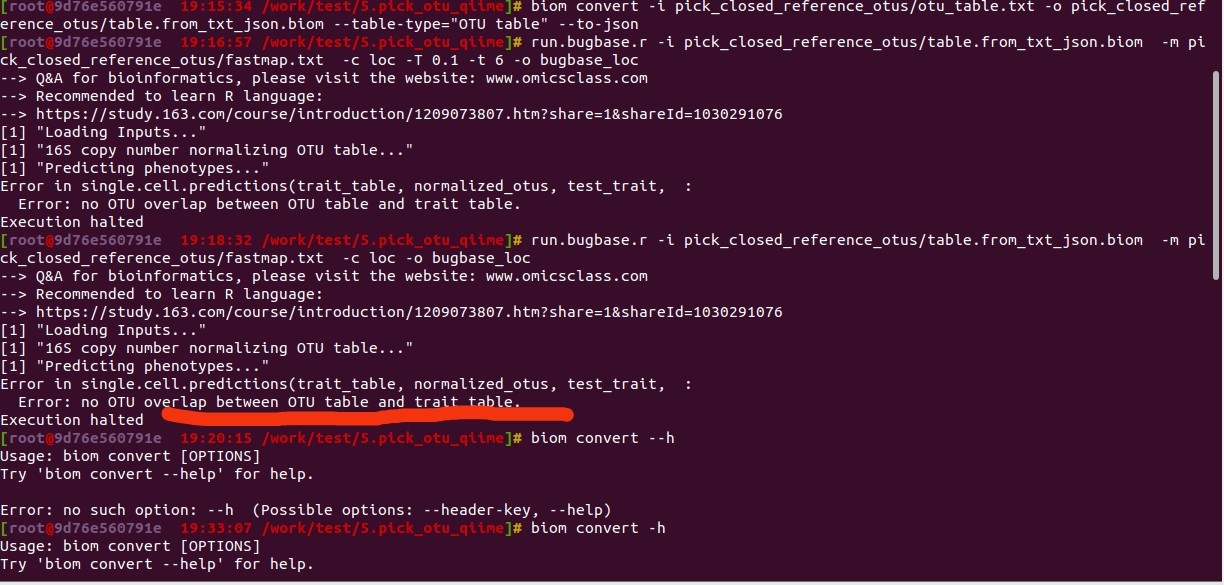
请先 登录 后评论
1 个回答
- 1 关注
- 0 收藏,2363 浏览
- Lucian 提出于 2023-08-12 20:24
