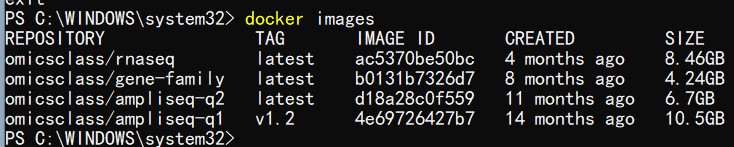
不知道你的电脑内存是多少?这一步比较消耗电脑内存;至少20G,或者30G以上的内存吧;
如果内存不够可能会报这样的错误;
Logging started at 07:29:00 on 17 Nov 2021
QIIME version: 1.9.1
qiime_config values:
blastmat_dir/biosoft/qiime-1.9.1/qiime_software/blast-2.2.22-release/data
pick_otus_reference_seqs_fp/biosoft/miniconda/envs/qiime1/lib/python2.7/site-packages/qiime_default_reference/gg_13_8_otus/rep_set/97_otus.fasta
sc_queueall.q
pynast_template_alignment_fp/biosoft/miniconda/envs/qiime1/lib/python2.7/site-packages/qiime_default_reference/gg_13_8_otus/rep_set_aligned/85_otus.pynast.fasta
cluster_jobs_fpstart_parallel_jobs.py
assign_taxonomy_reference_seqs_fp/biosoft/miniconda/envs/qiime1/lib/python2.7/site-packages/qiime_default_reference/gg_13_8_otus/rep_set/97_otus.fasta
torque_queuefriendlyq
jobs_to_start1
denoiser_min_per_core50
assign_taxonomy_id_to_taxonomy_fp/biosoft/miniconda/envs/qiime1/lib/python2.7/site-packages/qiime_default_reference/gg_13_8_otus/taxonomy/97_otu_taxonomy.txt
temp_dir/work/tmp/
blastall_fp/biosoft/qiime-1.9.1/qiime_software/blast-2.2.22-release/bin/blastall
seconds_to_sleep1
parameter file values:
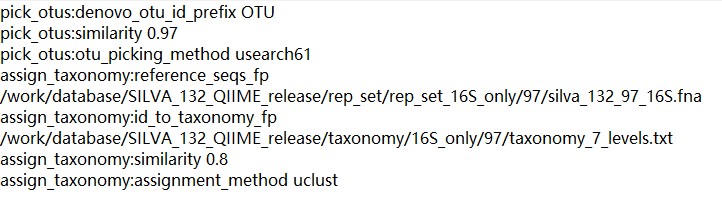
assign_taxonomy:reference_seqs_fp/work/database/SILVA_132_QIIME_release/rep_set/rep_set_16S_only/97/silva_132_97_16S.fna
assign_taxonomy:id_to_taxonomy_fp/work/database/SILVA_132_QIIME_release/taxonomy/16S_only/97/taxonomy_7_levels.txt
assign_taxonomy:assignment_methoduclust
assign_taxonomy:similarity0.8
parallel:jobs_to_start1
pick_otus:otu_picking_methodusearch61
pick_otus:denovo_otu_id_prefixOTU
pick_otus:similarity0.97
Input file md5 sums:
qiime.fasta: 898a5979a567ed66b9fad2008eb7b887
Executing commands.
# Pick OTUs command
pick_otus.py -i qiime.fasta -o pick_de_novo_otus/usearch61_picked_otus --otu_picking_method usearch61 --denovo_otu_id_prefix OTU --similarity 0.97
Stdout:
Stderr:
# Pick representative set command
pick_rep_set.py -i pick_de_novo_otus/usearch61_picked_otus/qiime_otus.txt -f qiime.fasta -l pick_de_novo_otus/rep_set//qiime_rep_set.log -o pick_de_novo_otus/rep_set//qiime_rep_set.fasta
Stdout:
Stderr:
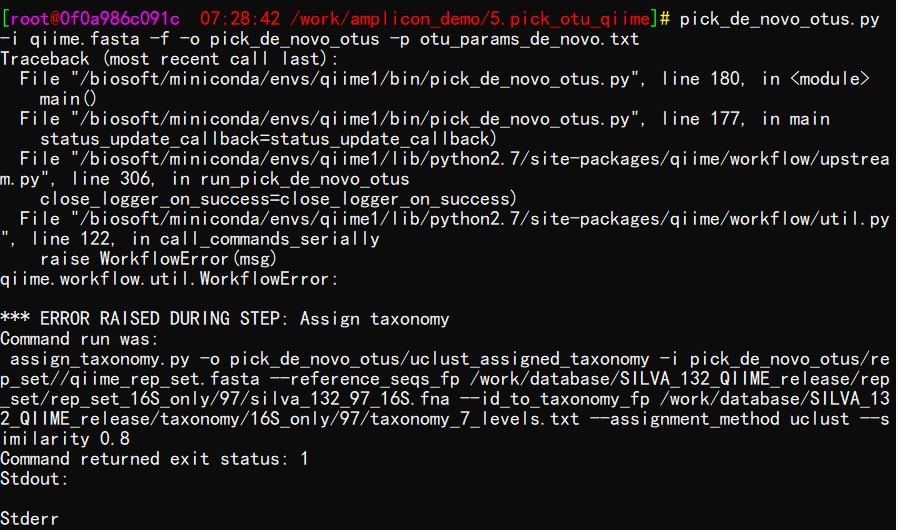
# Assign taxonomy command
assign_taxonomy.py -o pick_de_novo_otus/uclust_assigned_taxonomy -i pick_de_novo_otus/rep_set//qiime_rep_set.fasta --reference_seqs_fp /work/database/SILVA_132_QIIME_release/rep_set/rep_set_16S_only/97/silva_132_97_16S.fna --id_to_taxonomy_fp /work/database/SILVA_132_QIIME_release/taxonomy/16S_only/97/taxonomy_7_levels.txt --assignment_method uclust --similarity 0.8
*** ERROR RAISED DURING STEP: Assign taxonomy
Command run was:
assign_taxonomy.py -o pick_de_novo_otus/uclust_assigned_taxonomy -i pick_de_novo_otus/rep_set//qiime_rep_set.fasta --reference_seqs_fp /work/database/SILVA_132_QIIME_release/rep_set/rep_set_16S_only/97/silva_132_97_16S.fna --id_to_taxonomy_fp /work/database/SILVA_132_QIIME_release/taxonomy/16S_only/97/taxonomy_7_levels.txt --assignment_method uclust --similarity 0.8
Command returned exit status: 1
Stdout:
Stderr
sh: line 1: 403 Segmentation fault uclust --input "pick_de_novo_otus/rep_set//qiime_rep_set.fasta" --id 0.8 --rev --maxaccepts 3 --allhits --libonly --lib "/work/database/SILVA_132_QIIME_release/rep_set/rep_set_16S_only/97/silva_132_97_16S.fna" --uc "/work/tmp/UclustConsensusTaxonAssigner_Vl12U3.uc" > "/work/tmp/tmplRA108Az2t8hLynL08ju.txt" 2> "/work/tmp/tmpcgBTMr0lP5iSIzpLeTMX.txt"
Traceback (most recent call last):
File "/biosoft/miniconda/envs/qiime1/bin/assign_taxonomy.py", line 417, in <module>
main()
File "/biosoft/miniconda/envs/qiime1/bin/assign_taxonomy.py", line 394, in main
log_path=log_path)
File "/biosoft/miniconda/envs/qiime1/lib/python2.7/site-packages/qiime/assign_taxonomy.py", line 1304, in __call__
'--uc': uc_path})
File "/biosoft/miniconda/envs/qiime1/lib/python2.7/site-packages/burrito/util.py", line 285, in __call__
'StdErr:\n%s\n' % open(errfile).read())
burrito.util.ApplicationError: Unacceptable application exit status: 139
Command:
cd "/work/amplicon_demo/5.pick_otu_qiime/"; uclust --input "pick_de_novo_otus/rep_set//qiime_rep_set.fasta" --id 0.8 --rev --maxaccepts 3 --allhits --libonly --lib "/work/database/SILVA_132_QIIME_release/rep_set/rep_set_16S_only/97/silva_132_97_16S.fna" --uc "/work/tmp/UclustConsensusTaxonAssigner_Vl12U3.uc" > "/work/tmp/tmplRA108Az2t8hLynL08ju.txt" 2> "/work/tmp/tmpcgBTMr0lP5iSIzpLeTMX.txt"
StdOut:
StdErr:
Logging stopped at 07:37:55 on 17 Nov 2021