单细胞转录组数据挖掘流程记录-LUAD(GSE131907)
单细胞转录组数据挖掘流程记录-LUAD(GSE131907)
数据介绍:
来自谷歌翻译:
我们对 44 名患者中 58 例肺腺癌衍生的 208,506 个细胞进行了单细胞 RNA 测序(scRNA-seq),涵盖原发肿瘤、淋巴结和脑转移及胸腔积液,此外还包括正常肺组织和淋巴结。丰富的单细胞谱描绘了肺腺癌进展的复杂细胞图谱,包括周围肿瘤微环境中的癌症、间质和免疫细胞。
数据下载地址:https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131907
数据下载:
wget -c "https://ftp.ncbi.nlm.nih.gov/geo/series/GSE131nnn/GSE131907/suppl/GSE131907%5FLung%5FCancer%5Fraw%5FUMI%5Fmatrix.txt.gz" -O GSE131907_Lung_Cancer_raw_UMI_matrix.txt.gz wget -c "https://ftp.ncbi.nlm.nih.gov/geo/series/GSE131nnn/GSE131907/suppl/GSE131907%5FLung%5FCancer%5Fcell%5Fannotation.txt.gz" -O GSE131907_Lung_Cancer_cell_annotation.txt.gz
样本合并的,需要按样本分开方便后续度入质控,这里写了R代码:
#!/usr/bin/env Rscript
# ===============================
# 参数
# ===============================
anno_dir <- "./" # 存放 LUNG_N01.txt 等文件的目录
umi_file <- "GSE131907_Lung_Cancer_raw_UMI_matrix.txt.gz"
out_dir <- "./split"
dir.create(out_dir, showWarnings = FALSE)
# ===============================
# 读取 UMI 矩阵
# ===============================
message("Reading UMI matrix...")
umi <- read.table(
gzfile(umi_file),
header = TRUE,
sep = "\t",
check.names = FALSE,
stringsAsFactors = FALSE
)
gene_col <- umi[, 1, drop = FALSE]
umi_mat <- umi[, -1, drop = FALSE]
message("UMI matrix dimensions:")
print(dim(umi_mat))
# ===============================
# 处理每个 Sample
# ===============================
anno_files <- list.files(anno_dir, pattern = "\\.txt$", full.names = TRUE)
for (f in anno_files) {
sample_name <- sub("\\.txt$", "", basename(f))
message("Processing ", sample_name)
# 读取 annotation
anno <- read.table(
f,
header = TRUE,
sep = "\t",
stringsAsFactors = FALSE
)
idx <- anno$Index
idx <- intersect(idx, colnames(umi_mat))
if (length(idx) == 0) {
warning("No matching cells for ", sample_name)
next
}
sub_mat <- umi_mat[, idx, drop = FALSE]
out <- cbind(gene_col, sub_mat)
out_file <- file.path(out_dir, paste0(sample_name, "_UMI.txt.gz"))
write.table(
out,
gzfile(out_file),
sep = "\t",
quote = FALSE,
row.names = FALSE
)
message(" Saved: ", out_file,
" (", ncol(sub_mat), " cells)")
}
message("Done.")

细胞注释文件也分开:
zcat GSE131907_Lung_Cancer_cell_annotation.txt.gz | \
awk -F'\t' '
NR==1 {
header=$0
next
}
{
out=$3 ".txt"
if (!(out in seen)) {
print header > out
seen[out]=1
}
print >> out
}'
数据分析:
样本准备mata文件:meta.tsv
| Sample | Patient_id | Tissue | Histology | Smoking | Pathology | EGFR_MUT | EGFR_Type | Stages | Stages_Status |
| LUNG_N01 | P0001 | nLung | ADC | Never | MD | WT | WT | IA | Low |
| LUNG_N06 | P0006 | nLung | ADC | Ex | MD | NA | NA | IA | Low |
| LUNG_N08 | P0008 | nLung | ADC | Never | MD | L858R | MUT | IB | Low |
| LUNG_N09 | P0009 | nLung | ADC | Ex | PD | WT | WT | IIA | Low |
| LUNG_N18 | P0018 | nLung | ADC | Ex | MD | del19 | MUT | IA | Low |
| LUNG_N19 | P0019 | nLung | ADC | Cur | WD | exon_20 | MUT | IA | Low |
| LUNG_N20 | P0020 | nLung | ADC | Cur | PD | WT | WT | IA | Low |
| LUNG_N28 | P0028 | nLung | ADC(Double) | Cur | NA | WT | WT | IIIA | High |
| LUNG_N30 | P0030 | nLung | ADC | Never | NA | del19 | MUT | IA | Low |
| LUNG_N31 | P0031 | nLung | ADC | Ex | NA | WT | WT | IIIA | High |
| LUNG_N34 | P0034 | nLung | ADC | Never | MD | WT | WT | IA3 | Low |
| LUNG_T06 | P0006 | tLung | ADC | Ex | MD | NA | NA | IA | Low |
| LUNG_T08 | P0008 | tLung | ADC | Never | MD | L858R | MUT | IB | Low |
| LUNG_T09 | P0009 | tLung | ADC | Ex | PD | WT | WT | IIA | Low |
| LUNG_T18 | P0018 | tLung | ADC | Ex | MD | del19 | MUT | IA | Low |
| LUNG_T19 | P0019 | tLung | ADC | Cur | WD | exon_20 | MUT | IA | Low |
| LUNG_T20 | P0020 | tLung | ADC | Cur | PD | WT | WT | IA | Low |
| LUNG_T25 | P0025 | tLung | ADC(Double) | Ex | NA | WT | WT | IA | Low |
| LUNG_T28 | P0028 | tLung | ADC(Double) | Cur | NA | WT | WT | IIIA | High |
| LUNG_T30 | P0030 | tLung | ADC | Never | NA | del19 | MUT | IA | Low |
| LUNG_T31 | P0031 | tLung | ADC | Ex | NA | WT | WT | IIIA | High |
| LUNG_T34 | P0034 | tLung | ADC | Never | MD | WT | WT | IA3 | Low |
| EBUS_06 | P1006 | tL/B | ADC | Cur | PD | WT | WT | IV | High |
| EBUS_28 | P1028 | tL/B | ADC | Ex | NA | WT | WT | IV | High |
| EBUS_49 | P1049 | tL/B | ADC | Cur | PD | WT | WT | IV | High |
| BRONCHO_58 | P1058 | tL/B | ADC | Never | PD | NA | NA | IV | High |
| EBUS_10 | P1010 | mLN | ADC | Ex | NA | WT | WT | IV | High |
| BRONCHO_11 | P1011 | mLN | ADC | Never | NA | L858R | MUT | IV | High |
| EBUS_12 | P1012 | mLN | ADC | Never | NA | WT | WT | IV | High |
| EBUS_13 | P1013 | mLN | ADC | Cur | PD | WT | WT | IV | High |
| EBUS_15 | P1015 | mLN | ADC | Cur | NA | exon_18_(G719X)__exon_20_(S768I) | MUT | IIIA | High |
| EBUS_19 | P1019 | mLN | ADC | Never | NA | del19 | MUT | IV | High |
| EBUS_51 | P1051 | mLN | ADC | Ex | NA | WT | WT | IV | High |
| LN_01 | P2001 | nLN | ADC | Cur | PD | WT | WT | IIB | Low |
| LN_02 | P2002 | nLN | ADC | Never | MD | L858R | MUT | IB | Low |
| LN_03 | P2003 | nLN | ADC | Ex | MD | L858R | MUT | IIB | Low |
| LN_04 | P2004 | nLN | ADC | Cur | MD | L858R | MUT | IA2 | Low |
| LN_05 | P2005 | nLN | ADC | Never | MD | del19 | MUT | IA3 | Low |
| LN_06 | P2006 | nLN | ADC | Never | MD | NA | NA | IA3 | Low |
| LN_07 | P2007 | nLN | ADC | Ex | MD | WT | WT | IA | Low |
| LN_08 | P2008 | nLN | ADC | Never | MD | WT | WT | IB | Low |
| LN_11 | P2011 | nLN | ADC | Never | MD | L858R | MUT | IB | Low |
| LN_12 | P2012 | nLN | ADC | Cur | MD | WT | WT | IA | Low |
| EFFUSION_06 | P1006 | PE | ADC | Cur | PD | WT | WT | IV | High |
| EFFUSION_11 | P1011 | PE | ADC | Never | NA | L858R | MUT | IV | High |
| EFFUSION_12 | P1012 | PE | ADC | Never | NA | WT | WT | IV | High |
| EFFUSION_13 | P1013 | PE | ADC | Cur | PD | WT | WT | IV | High |
| EFFUSION_64 | P1064 | PE | ADC | Ex | NA | NA | NA | IV | High |
| NS_02 | P3002 | mBrain | ADC | Never | NA | WT | WT | IV | High |
| NS_03 | P3003 | mBrain | ADC | Never | NA | p.L858R | MUT | IV | High |
| NS_04 | P3004 | mBrain | ADC | Ex | NA | WT | WT | IV | High |
| NS_06 | P3006 | mBrain | ADC | Ex | PD | WT | WT | IV | High |
| NS_07 | P3007 | mBrain | ADC | Never | NA | WT | WT | IV | High |
| NS_12 | P3012 | mBrain | ADC | Never | NA | del19_L858R | MUT | IV | High |
| NS_13 | P3013 | mBrain | ADC | Ex | NA | G719S_S768I | MUT | IV | High |
| NS_16 | P3016 | mBrain | ADC | Cur | PD | WT | WT | IIIA | High |
| NS_17 | P3017 | mBrain | ADC | Never | NA | NA | NA | IV | High |
| NS_19 | P3019 | mBrain | ADC | Never | NA | NA | NA | IV | High |
这次的数据是 h5 格式的也可以直接读入:
cat ~/LUAD/data/meta.tsv | sed '1d' | \
parallel -j 10 --colsep '\t' '
Rscript $scripts/seurat_sc_qc.r \
--count ~/LUAD/data/split/{1}_UMI.txt.gz \
-p {1} --project {1} \
--nUMI.min 100 \
--nUMI.max 150000 \
--nGene.min 200 \
--nGene.max 10000 \
--mito.gene.pattern "^MT.*-" \
--percent_mito 20 \
--log10GenesPerUMI 0.8 \
--metadata ~/LUAD/data/{1}.txt \
--metadata.col.name Patient_id Tissue Histology Smoking Pathology EGFR_MUT EGFR_Type Stages Stages_Status \
--metadata.value {2} {3} {4} {5} {6} {7} {8} {9} {10}
'
#细胞周期和双细胞去除
cat ~/LUAD/data/meta.tsv|sed '1d'|while read Sample Patient_id Tissue Histology Smoking Pathology EGFR_MUT EGFR_Type Stages Stages_Status;do
Rscript $scripts/seurat_sc_cluster.r --rds $Sample.CellCycleScoring.qs \
--resolution 0.5 -d 30 \
-p $Sample -o $Sample --cpu 20
## 如果是 SCT 标准化需要加参数:--sct
## 如果要去除双细胞增加参数:--removeDoubletCells
Rscript $scripts/DoubletFinder.r -i $Sample/$Sample.qs \
-p $Sample --annotations seurat_clusters --removeDoubletCells
done
#合并样本,
Rscript $scripts/merge_seurat_obj.r -i .doubletFinder.qs -p all.sample.merged
# 分群聚类
Rscript $scripts/seurat_sc_cluster.r --cpu 10 --rds all.sample.merged.qs \
--integrate.method harmony --batch.id Sample \
--resolution 0.2 -d 50 \
-p luad.harmony -o luad.harmony
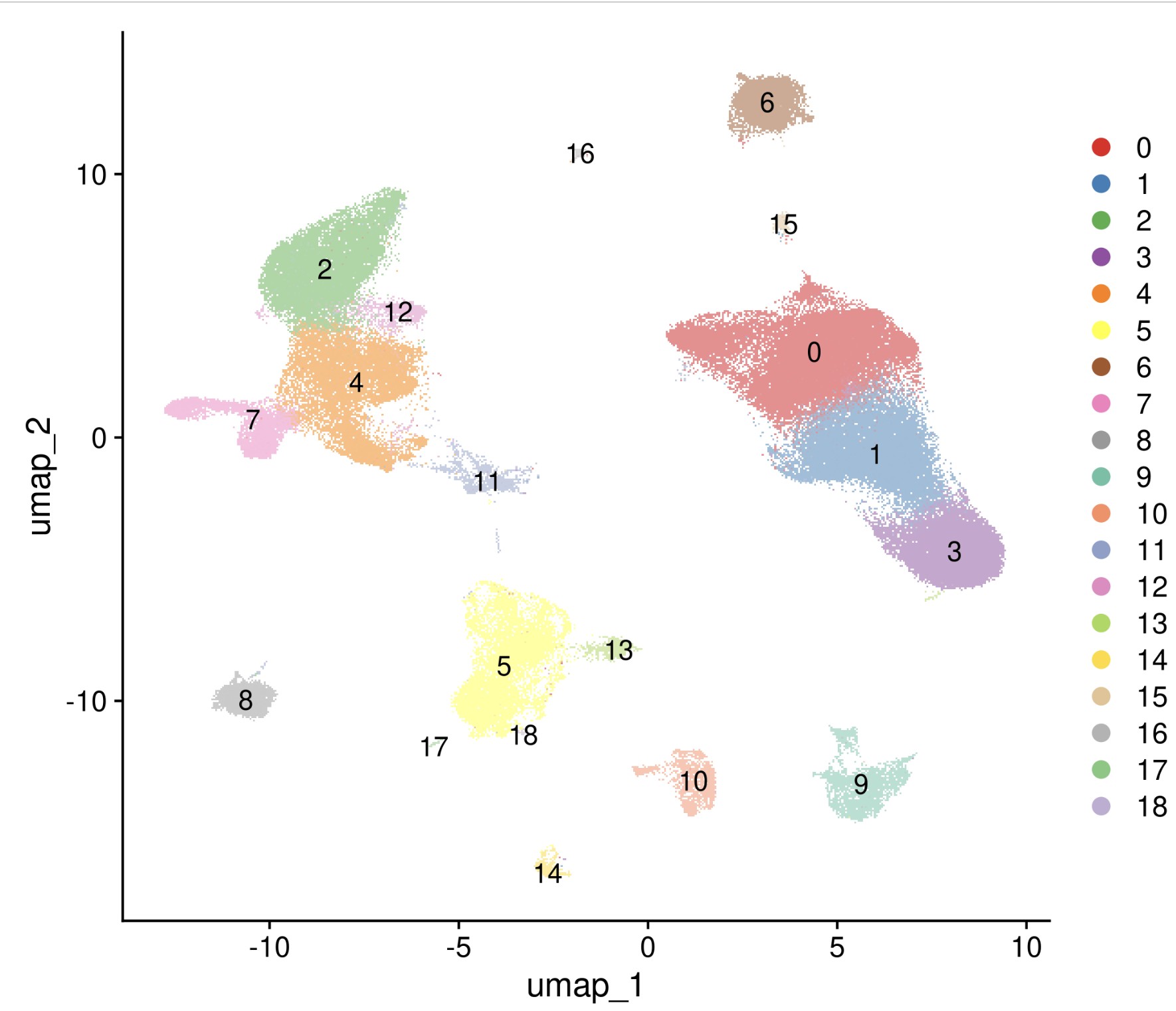
分析结果:
- 发表于 2026-01-13 14:05
- 阅读 ( 157 )
- 分类:转录组
你可能感兴趣的文章
相关问题
0 条评论
请先 登录 后评论
