network.r绘制网络图并输出最大子图节点及网络图
network.r绘制网络图,边权重信息决定网络中边的粗细,节点的度决定节点大小,根据节点类别填充节点颜色;提取网络的最大子图,输出构成最大子图的节点并绘制子图网络。
使用方法:
Rscript network.r -h
usage: network.r [-h] -e filepath -v filepath -f from -t to -w weight -g group
[--isolated] [-l style] [--label.node] [--sub_label.node]
[-o path] [-n prefix] [-H number] [-W number]
network plot:https://www.omicsclass.com/article/1531
optional arguments:
-h, --help show this help message and exit
-e filepath, --edge filepath
input the edge information of network,[required]
-v filepath, --vertex filepath
input the vertex information of network,[required]
-f from, --from from the name of the start gene column [required]
-t to, --to to the name of the end gene column[required]
-w weight, --combined_score weight
the name of the edge weight column[required]
-g group, --group group
input group id in vertex file, can set multi
groups[required]
--isolated delete isolated vertex [optional, default: False]
-l style, --layout style
set Network display style,c('star', 'circle', 'gem',
'dh', 'graphopt', 'grid', 'mds', 'randomly', 'fr',
'kk', 'drl', 'lgl'),[default fr]
--label.node show node name [optional, default: TRUE]
--sub_label.node show node name of sub-network [optional, default:
TRUE]
-o path, --outdir path
output file directory [default cwd]
-n prefix, --name prefix
out file name prefix [default demo]
-H number, --height number
the height of pic inches [default 12]
-W number, --width number
the width of pic inches [default 12]
参数说明:
-e 网络图的边信息文件路径,文件中包含边的起始节点,结尾节点,以及边的权重信息
| from | to | combined_score | fromGeneName | toGeneName |
| 9606.ENSP00000011653 | 9606.ENSP00000046794 | 959 | CD4 | LCP2 |
| 9606.ENSP00000061240 | 9606.ENSP00000204604 | 952 | TLL1 | CHRD |
| 9606.ENSP00000172229 | 9606.ENSP00000228945 | 945 | NGFR | ARHGDIB |
-v 网络中节点信息文件路径,文件中包含节点(节点名与边信息文件中起始节点名,结尾节点名相对应)和节点类别信息
| gene | regulate |
| CD4 | Up |
| TLL1 | Up |
| ARHGDIB | Up |
-f 边信息文件中起始节点列名
-t 边信息文件中结尾节点列名
-w 边信息文件中边权重列名
-g 节点信息文件中节点类别列名
-i 网络图中是否隐藏孤立节点,默认不隐藏
-L 是否标注完整网络的节点名,默认标注
-S 是否标注最大子图中节点名,默认标注
使用举例:
结果展示:
完整网络图
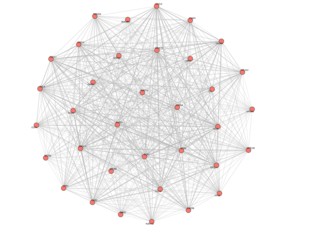
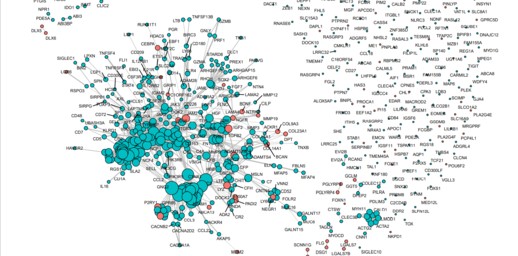 最大子图:
最大子图:
- 发表于 2021-08-23 11:22
- 阅读 ( 4525 )
- 分类:科研作图
你可能感兴趣的文章
0 条评论
请先 登录 后评论
