使用方法:
Rscript /share/nas1/zhangll/MAF/code/maf_oncoplot.r -h
usage: /share/nas1/zhangll/MAF/code/maf_oncoplot.r [-h] -i maffile -m metadata
-g group
[-a additive [additive ...]]
[-T topgene]
[-l genelist [genelist ...]]
[-t] [-o outdir]
[-H height] [-W width]
Mutation annotation file classification
visualization:https://www.omicsclass.com/article/1518
optional arguments:
-h, --help show this help message and exit
-i maffile, --maf maffile
input the maf file[required]
-m metadata, --meta metadata
input metadata file path[required]
-g group, --group group
input group id in metadata file to classification
visualization[required]
-a additive [additive ...], --additive additive [additive ...]
add additional class[optional,default: NULL]
-T topgene, --top topgene
Number of top genes displayed [optional, default: 20]
-l genelist [genelist ...], --genelist genelist [genelist ...]
the selected gene list to display [optional, default:
NULL]
-t, --showtitle whether show maftools title of plot [optional,
default: False]
-o outdir, --outdir outdir
output file directory [default cwd]
-H height, --height height
the height of pic inches [default 8]
-W width, --width width
the width of pic inches [default 8]
参数说明:
-i 突变注释文件maf的路径,可从TCGA数据库中直接下载:
| | | | | | | | |
| | | GRCh38 | | | 12001777
| | |
| | | GRCh38 | | | 42444578
| + | Missense_Mutation |
| | WUGSC | GRCh38
| | 209692762
| 209692762 | | |
| | | GRCh38
| | | 125010546 | | |
| | | GRCh38
| | | 141200541
| | |
| | | GRCh38
| | | 177294257
| | |
| | WUGSC | GRCh38
| | | 6174609
| | |
| | | GRCh38
| | | 128359624
| | |
| | | GRCh38
| | | 94558941
| | |
-m metadata文件,病人样本的各种特征信息,必须包含以patient命名的患者TCGA编号列:
| | | | tissue_or_organ_of_origin
| | | | |
| | | | | | | | |
| | | | | | | | |
| | | | Middle third of esophagus
| | | | |
| | | | | | | | |
| | | | | | | | |
| | | | | | | | |
| | | | | Middle third of esophagus |
| | | | |
| | | | | Middle third of esophagus |
| | | | |
-g 指定metadata 分组列名,如果分组名字有空格,应该用引号引起来
-a 附加分组列名,结果将在图中显示
-T 可指定所要展示的top基因个数,默认20
-l 可指定所要展示的基因列表,基因之间用空格隔开,默认NULL
使用举例:
Rscript /share/nas1/zhangll/MAF/code/maf_oncoplot.r -i /share/nas1/zhangll/MAF/TCGA-ESCA.maf_maftools.maf -m /share/nas1/huangls/project/TCGA_ESCA/metadata.tsv -g gender
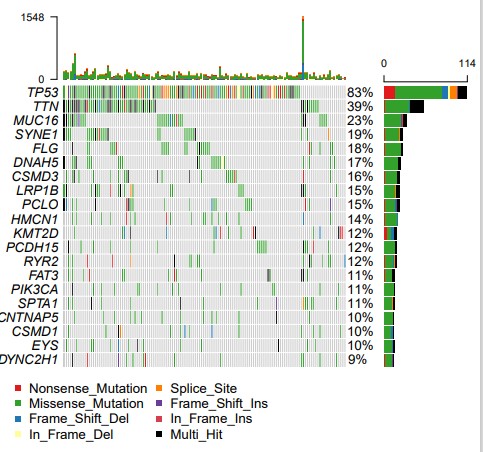
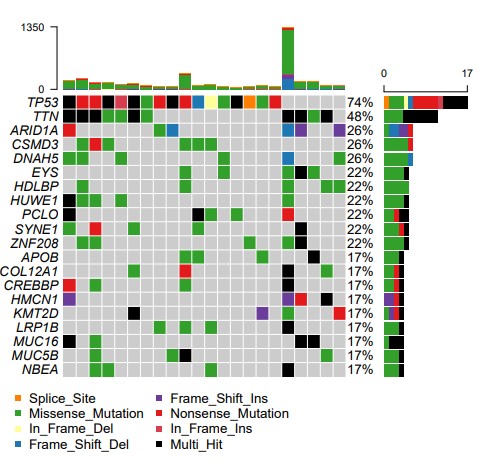
结果展示:
瀑布图: