怀疑你的路径写错错了,你检查一下:
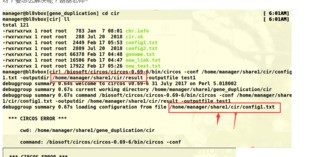
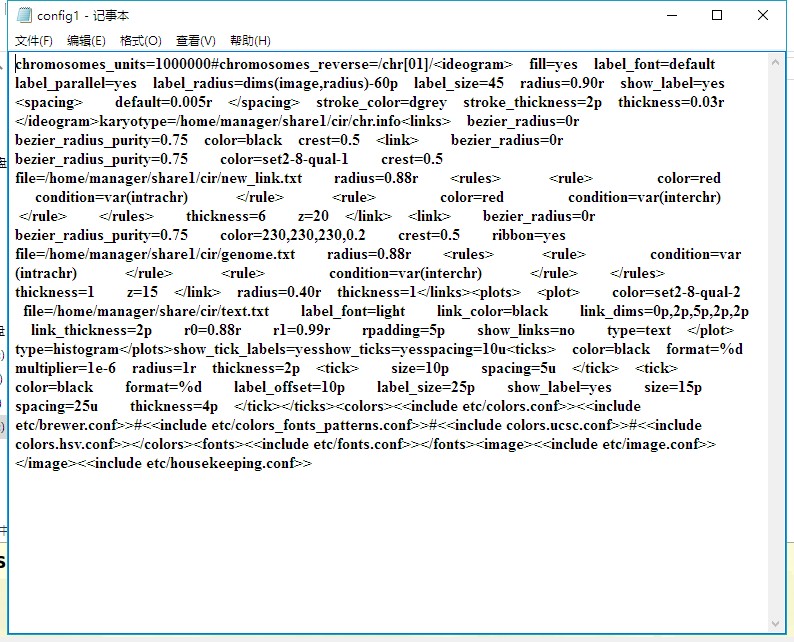
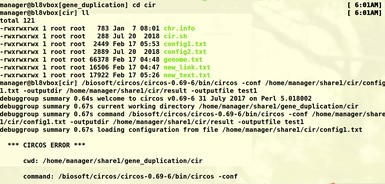
老师您好!根据老师前几日给出的建议采用circos进行基因组共线性分析。link,text,genome文件按照老师课程中的格式准备。config1.txt 用的是老师给的参考资料中的文件,仅修改了输入文件。最后运行绘图命令时出现如下报错,请问老师这个是哪里出问题了呀?要怎么解决呢?谢谢老师~
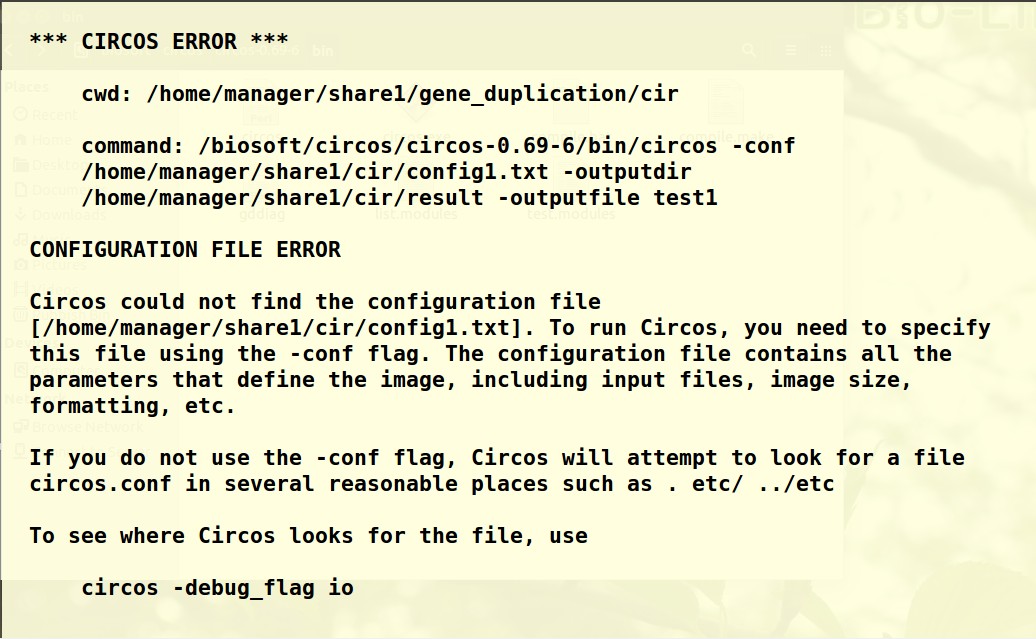
To see where Circos looks for the file, use
circos -debug_flag io
To see how configuration files work, create the example image, whose
configuration and data are found in example/. From the Circos distribution
directory,
cd example
../bin/circos -conf ./circos.conf
or use the 'run' script (UNIX only).
Configuration files are described here
http://circos.ca/tutorials/lessons/configuration/configuration_files/
and the use of command-line flags, such as -conf, is described here
http://circos.ca/tutorials/lessons/configuration/runtime_parameters/
Windows users unfamiliar with Perl should read
http://circos.ca/tutorials/lessons/configuration/unix_vs_windows/
This error can also be produced if supporting configuration files, such as
track defaults, cannot be read.
If you are having trouble debugging this error, first read the best practices
tutorial for helpful tips that address many common problems
http://www.circos.ca/documentation/tutorials/reference/best_practices
The debugging facility is helpful to figure out what's happening under the
hood
http://www.circos.ca/documentation/tutorials/configuration/debugging
If you're still stumped, get support in the Circos Google Group.
http://groups.google.com/group/circos-data-visualization
Please include this error, all your configuration, data files and the version
of Circos you're running (circos -v).Do not email me directly -- please use
the group.
Stack trace:
at /biosoft/circos/circos-0.69-6/bin/../lib/Circos/Error.pm line 425.
Circos::Error::fatal_error('configuration', 'missing', '/home/manager/share1/cir/config1.txt') called at /biosoft/circos/circos-0.69-6/bin/../lib/Circos/Configuration.pm line 796
Circos::Configuration::loadconfiguration('/home/manager/share1/cir/config1.txt') called at /biosoft/circos/circos-0.69-6/bin/../lib/Circos.pm line 148
Circos::run('Circos', 'outputdir', '/home/manager/share1/cir/result', '_cwd', '/home/manager/share1/gene_duplication/cir', '_argv', '-conf /home/manager/share1/cir/config1.txt -outputdir /home/m...', 'outputfile', 'test1', ...) called at /biosoft/circos/circos-0.69-6/bin/circos line 536